RE-1 silencing transcription factor (REST) regulates human
synaptophysin gene transcription through an intronic
sequence-specific DNA-binding site
Michael Lietz, Mathias Hohl and Gerald Thiel
Department of Medical Biochemistry and Molecular Biology, University of Saarland Medical Center, Homburg, Germany
Synaptophysin, one of the major proteins on synaptic vesi-
cles, is ubiquitously expressed throughout the brain. Syn-
aptophysin and synapsin I, another synaptic vesicle protein,
are also expressed by retinoic acid-induced neuronally dif-
ferentiated P19 teratocarcinoma cells. Here, we show that
inhibition of histone deacetylase activity in P19 cells is suf-
ficient to activate transcription of the synaptophysin and
synapsin I genes, indicating that neuronal differentiation
and impairment of histone deacetylases results in a similar
gene expression pattern. The transcription factor REST, a
repressor of neuronal genes in non-neuronal tissues, has
been shown to function via recruitment of histone deacety-
lases to the transcription unit, indicating that modulation of
the chromatin structure via histone deacetylation is of major
importance for REST function and neuron-specific gene
transcription. Furthermore, REST has been shown to be the
major regulator of neuronal expression of synapsin I. Here,
we have identified a functional binding site for REST in the
first intron of the human synaptophysin gene indicating that
REST blocks human synaptophysin gene transcription
through an intronic neuron-specific silencer element. The
synaptophysin promoter is, however, devoid of neuron-
specific genetic elements and directs transcription in both
neuronal and non-neuronal cells. Using a dominant-negat-
ive approach we have identified the transcription factor Sp1
as one of the regulators responsible for constitutive tran-
scription of the human synaptophysin gene.
Keywords:neuronalgenes;REST;Sp1;synapsin I;synapto-
physin.
Synaptophysin is a major integral membrane protein of
small synaptic vesicles [1]. Synaptophysin forms a hetero-
multimeric complex in the vesicle membrane, consisting of
at least four synaptophysin molecules and the synaptic
vesicle protein synaptobrevin [2]. Many functions have been
attributed to synaptophysin in the past: synaptophysin has
been proposed to form a channel/fusion pore [3], to function
as a Ca
2+
sensor in the synapse [4], and to be essential for
neurotransmitter release [5]. Gene targeting experiments in
transgenic mice revealed, however, that synaptophysin is
not essential for neurotransmitter release. No difference
between wild-type and mutant mice in synaptic transmission
and short-term and long-term plasticity was observed [6]
suggesting that other synaptic vesicle proteins may com-
pensate for the lack of synaptophysin. Double knockout
mice lacking synaptophysin and the structurally related
synaptic vesicle protein synaptogyrin I showed a severe
reduction in short-term and long-term plasticity, indicating
that synaptophysin and synaptogyrin I perform essential,
redundant functions in synaptic plasticity [7]. Recently, a
further role of synaptophysin in regulating activity-depend-
ent synapse formation has been proposed [8].
Synaptophysin is ubiquitously expressed in neurons
throughout the brain and also in neuroendocrine cells [9].
During neuronal development, synaptophysin expression is
correlated with synaptogenesis [10] and synaptophysin has
been widely used as marker for neurons and nerve terminal
differentiation, due to its abundance and pan-neuronal
expression.
Here, we have analyzed the regulation of the human
synaptophysin gene, in comparison with the regulation of
the synapsin I gene. We show that expression of both genes
is sensitive to histone deacetylase inhibition, indicating that
chromatin structure governs synaptophysin and synapsin I
gene transcription. Furthermore, two transcription factors,
Sp1 and the RE-1 silencing transcription factor (REST)
were identified that are responsible for either constitutive or
neuron-specific transcription of the synaptophysin gene.
Experimental procedures
Reporter constructs
The reporter constructs are derivatives of pGL3-Basic
and pGL3-Promoter (Promega). To construct plasmid
pSyI
-2309/+47
luc we cut plasmid pSyCAT10 [11] with SalI
and ligated the fragment into the XhoIsiteofpGL3-Basic.
The genomic clone p10C-6 containing the promoter, exons I–
III and introns I and II of the human synaptophysin gene
Correspondence to G. Thiel, Department of Medical Biochemistry
and Molecular Biology, Building 44, University of Saarland
Medical Center, D-66421 Homburg, Germany.
Fax: + 49 6841 1626500, Tel.: + 49 6841 1622606,
E-mail: bcgthi@uniklinik-saarland.de
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase;
GST, glutathione S-transferase; NRSE, neural-restrictive silencer
element; REST, RE-1 silencing transcription factor; Syp,
synaptophysin; TSA, trichostatin A.
(Received 8 August 2002, revised 6 November 2002,
accepted 11 November 2002)
Eur. J. Biochem. 270, 2–9 (2003) FEBS 2003 doi:10.1046/j.1432-1033.2003.03360.x

[12], was a kind gift of T. Su
¨dhof, HHMI, Dallas, TX, USA.
To construct a synaptophysin promoter/luciferase reporter
gene, we first subcloned a KpnI fragment of the genomic
clone p10C-6 into pGEM7 (Promega). This plasmid was
modified by inserting the annealed oligonucleotides
5¢-CTAGCCGAGCCTCCCGCCCCCTGCATTGCTGG
TCGACGCATG-3¢and 5¢-CGTCGACCAGCAATGCA
GGGGGCGGGAGGCTCGG-3¢into the NheIandSphI
sites. This plasmid was subsequently cut with XbaI, filled in
with the Klenow fragment of Escherichia coli DNA poly-
merase I, and recut with SalI. The fragment encompassing the
5¢-flanking region of the human synaptophysin gene (nucle-
otides 29255–31636, accession no. U93305) was isolated and
inserted into plasmid pGL3-Basic, generating plasmid pSyp
-
2356/+27
luc. Plasmid pSyp
Intron
luc, containing nucleotides 3–
427 of the first intron of the human synaptophysin gene
(nucleotides 28788–29216, accession no. U93305) upstream
of the SV40 promoter, was constructed by inserting an RsaI
fragment derived from plasmid p10C-6 into Ecl136II-cut
pGL3-Promoter. Plasmids pSypNRSE
2
SV40luc and pSyI-
NRSE
2
SV40luc, containing two copies of the intronic NRSE
(neural-restrictive silencer element) derived from the synapt-
ophysin gene or two copies of the NRSE derived from the
human synapsin I promoter 5¢of the SV40 promoter, were
generated by subcloning of the synthetic oligonucleotides
5¢-TCGAGTCCAGCACCGTGGACAGAG CCG-3¢and
5¢-TCGACGGCTCTGTCCACGGTGCTG GAC-3¢(syn-
aptophysin gene) or 5¢-TCGAGCTTCAG CACCGCGG
ACAGTGCCTTG-3¢and 5¢-TCGACAA GGCACTGTC
CGCGGTGCTGAAGC-3¢(synapsin I gene) into the XhoI
and SalI sites of plasmid pHIVTATA CAT [13]. The
sequences were subsequently multimerized as described
previously [13], excised with XhoIandSalI and cloned into
the XhoI site of plasmid pGL3-Promoter.
Expression constructs
The REST expression vector pCMVFLAG-REST is iden-
tical to the previously described plasmid pCMVmycREST
[14], except that the myc epitope has been exchanged
for a triple FLAG epitope (sequence: MDYKDHDG
DYKDHDLDYKDDDDK). The expression vector enco-
ding a positive-dominant mutant of REST, FLAG-DP-
REST, is identical to the previously described plasmid
pCMVDP-REST [15,16], except that the hemagglutinin
epitope has been exchanged for a triple FLAG epitope. The
expression vector encoding myc-tagged REST4 [16] and
mammalian glutathione S-transferase (GST) encoding
expression vectors pEBGN and pEBGN-Sp1 [17,18] have
been described previously. Plasmids pRSVband pSV40lacZ
encodes for b-galactosidase of E. coli [19,20].
Cell culture and transfections
The murine neuroblastoma cell line NS20Y, the immortal-
ized septal cell line SN56, the murine teratocarcinoma cell
line P19 and human 293T cells were maintained as described
previously [11,19,21,22]. NS20Y cells were transfected using
the calcium phosphate coprecipitation method with 0.5–2 lg
of luciferase reporter plasmid, 100 ng of expression vector
encoding FLAG-REST, FLAG-DP-REST or myc-REST4,
and 0.5–1 lgofpRSVbinternal standard plasmid. 293T cells
were transfected as described previously [22] with 1 lgof
luciferase reporter plasmid, 1–5 lg of GST expression vector
and 0.2 lg of the internal standard plasmid pSV40lacZ. The
titration experiments with NS20Y cells were performed with
1lg of luciferase reporter plasmid, 1–5 lg of GST-expres-
sion vector and 0.2 lg of the internal standard plasmid
pSV40lacZ. P19 cells were grown as described previously
[23]. For neuronal differentiation, cells were aggregated and
treated with 0.5 l
M
all-trans-retinoid acid for 4 days. Cell
aggregates were then lightly trypsinized, plated onto tissue
culture plates and cultured for 5 days in the absence of
retinoid acid, but in the presence of 5 lgÆmL
)1
of cytosine
b-
D
-arabinofuranoside (Sigma C6645). Trichostatin A
(TSA) was purchased from Wako Chemicals GmbH (Neuss,
Germany) and used at a concentration of 100 ngÆmL
)1
dissolved in dimethylsulfoxide.
RNase protection mapping
Cytoplasmic RNA of undifferentiated, differentiated,
dimethylsulfoxide and TSA-treated P19 cells was prepared
as described previously [23]. For RNase protection mapping,
20 lg RNA was used for the detection of synapsin I and
synaptophysin mRNA, and 2.5 lg RNA for the detection of
glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
and b-actin mRNA. The template for mouse synapsin I
cRNA synthesis (plasmid pSP6-mSyI-2) has been described
previously [23]. To synthesize a synaptophysin-specific
riboprobe, a cDNA fragment encompassing nucleotides
1730–2011 of the mouse synaptophysin gene (accession no.
X95818) was amplified by PCR using the primers 5¢-
GGTTCTGGTCAGGATTGC-3¢and 5¢-TCAGTAAGG
GACATTTCG-3¢and cloned into the SmaIsiteofpBlue-
script to generate plasmid pT3mp38. Hybridization with
synapsin I and synaptophysin mRNA protected fragments
of 194 and 282 nucleotides, respectively, from RNase
digestion. Plasmids SP6-b-actin and pTRI-GAPDH-Rat,
used to synthesize b-actin and GAPDH-specific riboprobes,
were purchased from Ambion. Hybridization with b-actin
and GAPDH mRNA protected fragments of 250 and 316
nucleotides, respectively, from RNase digestion.
Reporter gene assays
Cell lysates of transfected cells were prepared 48 h post-
transfection using the cell culture lysis buffer (Promega), and
b-galactosidase and luciferase activities were determined as
described [20,21]. Each experiment included four separate
transfections for each experimental setting, and the experi-
ments were repeated at least twice giving consistent results.
Expression and purification of recombinant
GST fusion proteins
293T cells were transfected with plasmids pEBGN or
pEBGN-Sp1, encoding a nuclear targeted GST or a GST-
Sp1 fusion protein. Forty-eight hours post-transfection, cells
were harvested, washed with NaCl/P
i
and lyzed in RIPA
buffer (50 m
M
Tris/HCl, pH 7.5, 150 m
M
NaCl, 1 m
M
EDTA, 10% NP-40, 0.5% deoxycholate, 0.1% SDS) for
FEBS 2003 Regulation of synaptophysin gene transcription (Eur. J. Biochem. 270)3
30 min at 4 C and centrifuged for 10 min. The supernatant
was incubated with 50 lL glutathione-agaraose beads
(Pharmacia Biotech) for 30 min. The glutathione-agarose-
GST protein complexes were isolated by centrifugation,
washed twice with RIPA buffer and dissolved in 100 l
M
of
SDS-stop solution (125 m
M
Tris/HCl, pH 6.8, 3 m
M
EDTA, 20% glycerol, 9% SDS, 0.05% bromophenol blue).
Results
Induction of neuronal gene transcription in P19
teratocarcinoma cells by retinoid acid or TSA
The P19 teratocarcinoma cell line is frequently used as an
in vitro model system for neuronal differentiation, neurite
outgrowth and neuronal gene expression. We differentiated
P19 cells on bacterial plates in the presence of retinoid acid.
Four days later, the cell aggregates were lightly trypsinized
and plated onto tissue culture plates for 5 days in the
presence of cytosine b-
D
-arabinofuranoside to suppress
growth of non-neuronal cells. Cytoplasmic RNA was
prepared and analyzed by RNase protection mapping using
specific riboprobes for the detection of synapsin I, synapto-
physin, b-actin and GAPDH mRNA, respectively. Synap-
sin I and synaptophysin are synaptic vesicle proteins and
serve as marker proteins for neuronal differentiation. b-Actin
and GAPDH are constitutively expressed in P19 cells.
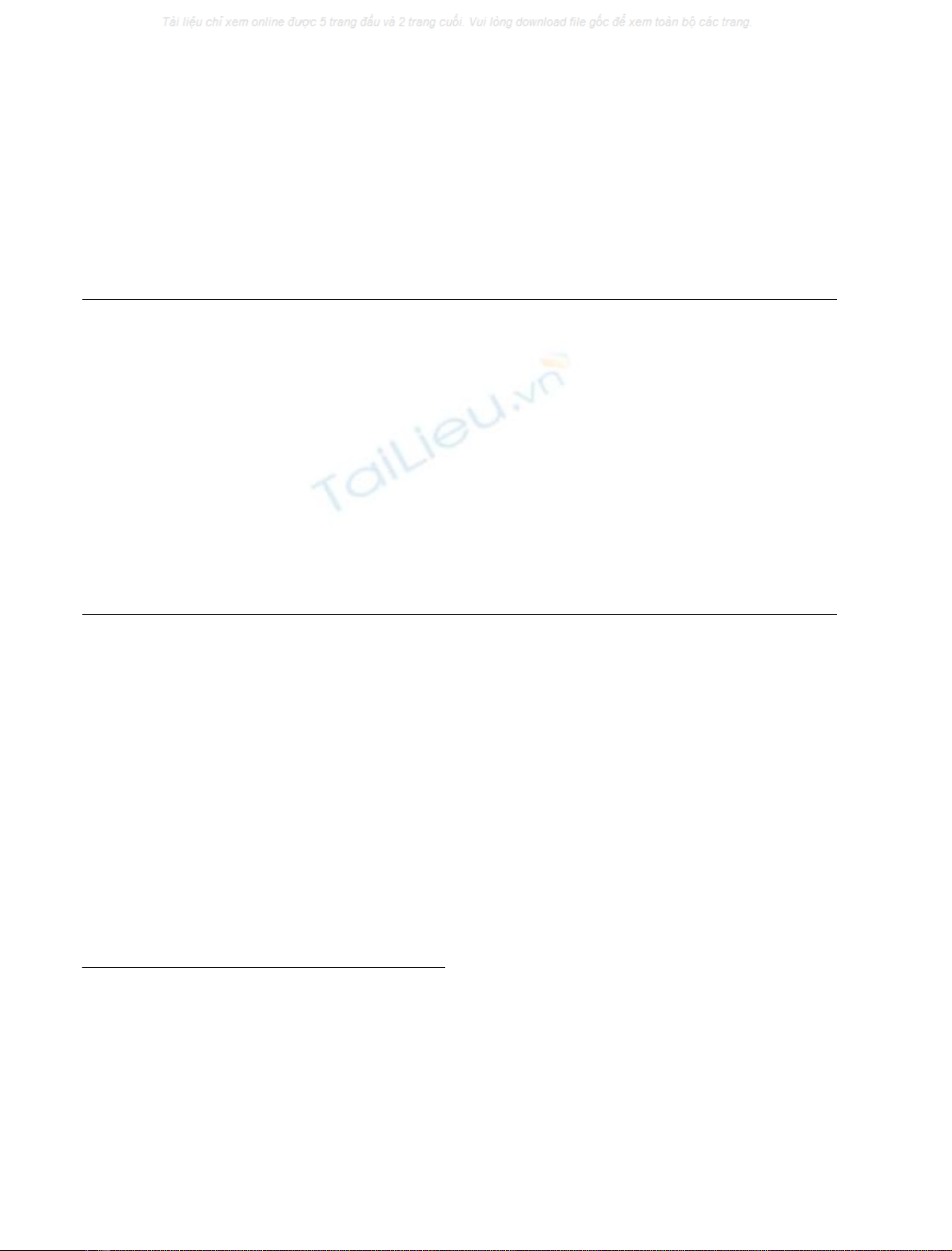
Figure 1A shows that neuronal differentiation of P19 cells
induced the expression of the synapsin I and synaptophysin
genes, confirming previous results [23,24]. Neuronal expres-
sion of the synapsin I gene has been shown to be controlled
by REST [25], a transcriptional repressor of neuronal genes
in non-neuronal cells. REST contains two activetranscrip-
tional repression domains on its N- and C-termini. The
N-terminal repression domain of REST recruits histone
deacetylases to the target genes of REST [26,27]. Likewise,
repression mediated by this domain was shown to be sensitive
to inhibitors of histone deacetylases such as TSA. Moreover,
there are indications that the C-terminal repression domain
also functions via recruitment of histone deacetylases [28].
Histone deacetylation generates a compact chromatin struc-
ture that is not as accessible to the transcriptional machinery.
Thus, alterations of the chromatin structure are essential for
transcriptional repression via REST. Therefore, we tested
whether an inhibition of histone deacetylases in P19 cells is
sufficient to induce neuronal gene transcription. P19 cells
weretreatedfor24hwithTSAandcytoplasmicRNAwas
prepared and analyzed by RNase protection mapping.
Figure 1B shows that inhibition of histone deacetylases by
TSA was sufficient to induce synapsin I gene transcription.
Moreover, TSA treatment also induced transcription of the
synaptophysin gene, indicating that expression of both genes
is controlled by alterations of the chromatin structure.
The human synaptophysin promoter is devoid
of neuron-specific genetic elements
The synapsin I promoter contains a neuron-specific control
element, the REST binding site termed neuron-restrictive
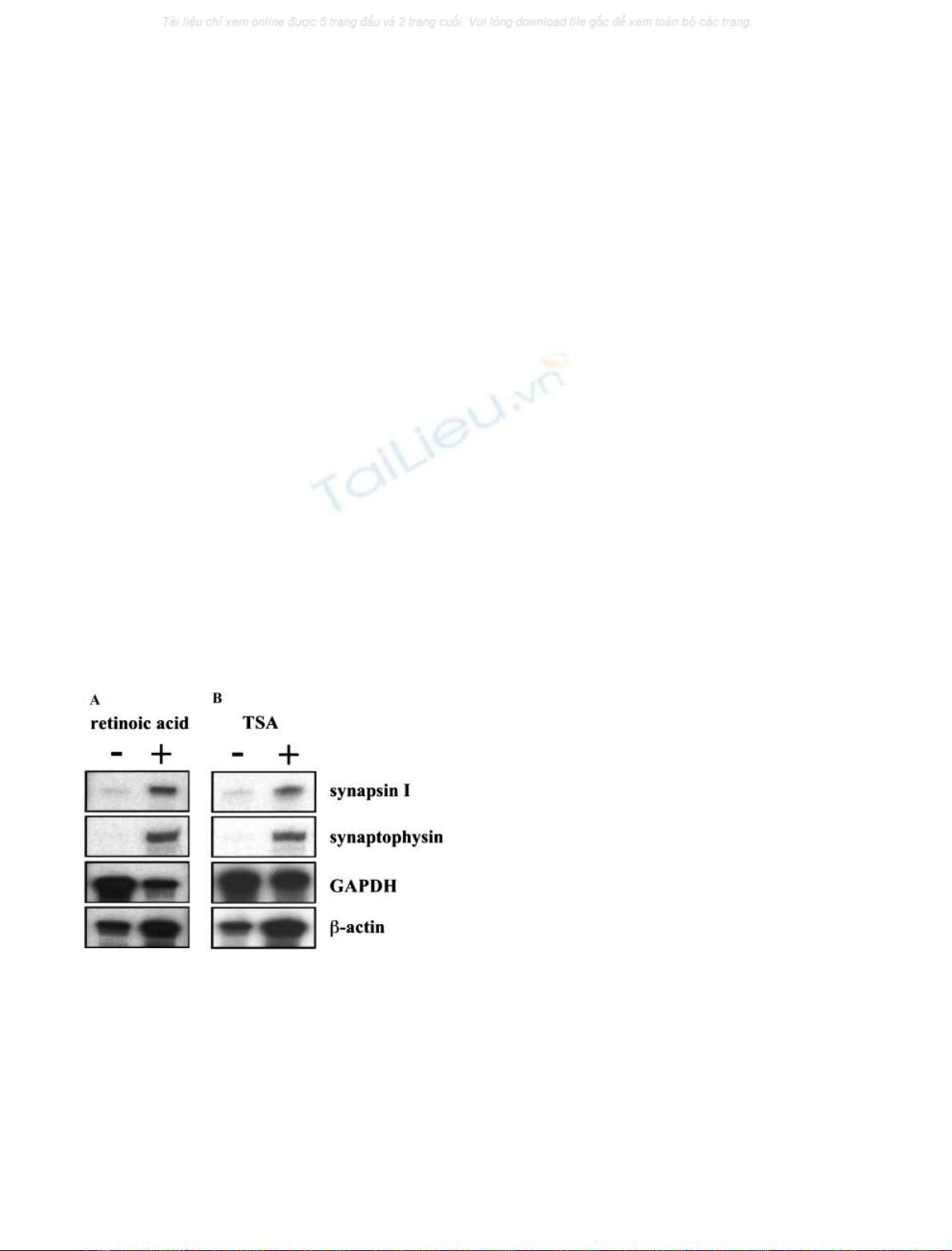
silencer element (NRSE). Transfection of a human synap-
sin I promoter/luciferase reporter gene depicted in Fig. 2A
in neuronal and non-neuronal cells revealed that the
promoter mainly directs luciferase expression in the neur-
onal cell lines NS20Y and SN56, but not in the human
embryonic kidney cell line 293T (Fig. 2B, upper panel).
Both NS20Y and SN56 cells have been shown by RNase
protection mapping to express synapsin I and synaptophy-
sin (data not shown). A human synaptophysin promoter/
luciferase reporter gene, however, showed constitutive
transcriptional activity in NS20Y, SN56 and 293T cells,
indicating that neuron-specific expression of synaptophysin
is not regulated by genetic elements located in the
5¢-flanking region of the synaptophysin gene.
Identification of a REST consensus binding site within
the first intron of the human synaptophysin gene
A data base search revealed the presence of an NRSE in the
first intron of the synaptophysin gene of the rat [29]. We
analyzed the database of the human genome and found an
NRSE in the first intron of the synaptophysin gene at an
identical position as in synaptophysin gene of the rat
(Fig. 3A). A comparison of the sequence revealed that the
intronic NRSE of the synaptophysin gene is 100%
conserved between the rat and human gene and has only
three mismatches in comparison to the NRSE found in the
human synapsin I promoter (Fig. 3B).
The intronic REST binding site confers REST regulation
to reporter genes
REST has been shown to repress transcription despite the
location or orientation of its binding site within a gene [14].
Fig. 1. Activation of synapsin I and synaptophysin gene transcription in
P19 teratocarcinoma cells. (A) P19 teratocarcinoma cells were differ-
entiated via aggregation and treatment with retinoic acid for 4 days,
than plated and cultured for a further 5 days in the presence of cytosine
b-
D
-arabinofuranoside. (B) P19 cells were treated for 24 h with the
histone deactylase inhibitor TSA or with the vehicle dimethylsulfoxide.
Cytoplasmic RNA from undifferentiated (A, denoted –), neuronally
differentiated (A, denoted +), dimethylsulfoxide-treated (B, denoted
–) and TSA-treated (B, denoted +) P19 cells were isolated and
analyzed by RNase protection mapping using cRNAs specific for
synapsin I, synaptophysin, b-actin and GAPDH, respectively.
4 M. Lietz et al. (Eur. J. Biochem. 270)FEBS 2003
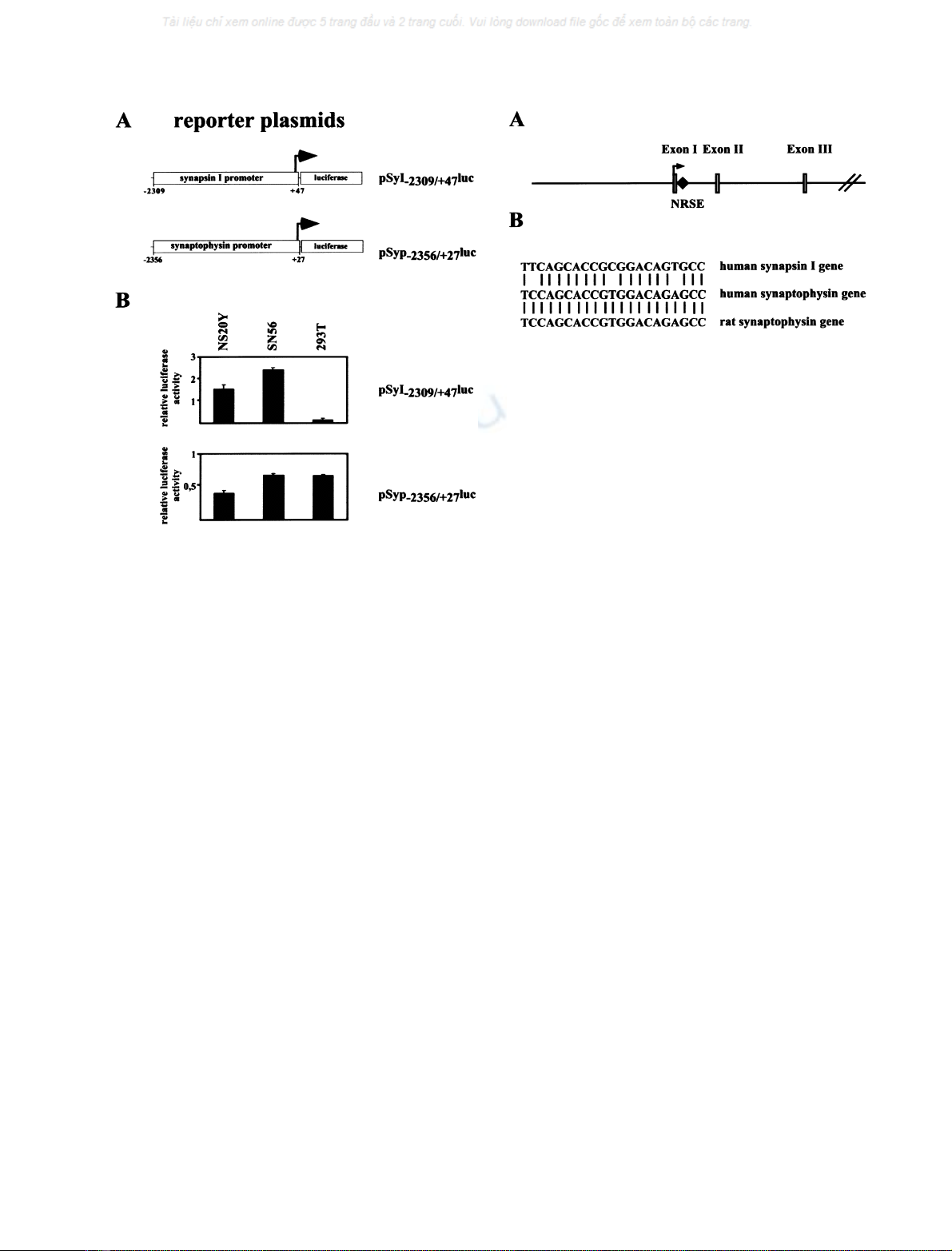
Thus, a functional NRSE of the synaptophysin gene should
operate from any position within the gene. We therefore
generated the reporter plasmid, pSyp
Intron
luc, consisting of
the luciferase open reading frame and the SV40 promoter.
To the 5¢of the SV40 promoter, we inserted 424 nucleotides
from the first intron of the human synaptophysin gene
including the NRSE. As a control, we used plasmid pGL3-
Promoter, containing the luciferase gene under control of
the SV40 promoter (Fig. 4A). As expression vectors, we
transfected plasmids encoding either a FLAG-tagged
REST, a FLAG-tagged DP-REST or a myc-tagged
REST4. The modular structure of FLAG-DP-REST and
myc-REST4, in comparison to REST, is depicted in
Fig. 4B. DP-REST contains the DNA-binding domain of
REST fused to the activation domain of the herpes simplex
virus protein VP16. Following binding to the REST cognate
site, DP-REST strongly activates transcription, due to the
presence of a transcriptional activation domain [15,16].
REST4 contains the N-terminal repression domain of
REST and five of the eight zinc fingers that constitutes the
DNA-binding domain [16]. REST4 binds only weakly to
the NRSE, due to the lack of zinc finger 7 that is important
for DNA binding [30]. REST4 was used as a negative
control in the experiment. Transient tranfections were
performed with NS20Y neuroblastoma cells. One of the
reporter plasmids, pSyp
Intron
luc or pGL3-Promoter, was
transfected into NS20Y cells together with plasmid pRSVb,
encoding b-galactosidase under the control of the Rous
sarcoma virus long-terminal repeat, to correct for variations
in transfection efficiencies. In addition, the emptyexpres-
sion vector pCMV5 (control) or expression vectors enco-
ding FLAG-REST, FLAG-DP-REST or myc-REST4 were
transfected. Forty-eight hours post-transfection, cells were
harvested, cell extracts prepared and the relative luciferase
activities determined. The results show that FLAG-REST
repressed transcription of the transcription unit containing
part of the first intron of the human synaptophysin gene
(plasmid pSyp
Intron
luc). Likewise, expression of FLAG-DP-
REST increased reporter gene expression over the level
already obtained by the strong SV40 promoter (Fig. 4C, left
panel). In contrast, myc-REST4 did not show any tran-
scriptional activity as expected from previous experiments
[16]. Naturally, the SV40 promoter was not regulated by
either FLAG-REST, FLAG-DP-REST or myc-REST4
(Fig. 4C, right panel). These data indicate that the NRSE
derived from the human synaptophysin gene is biological
active and functions as a REST-regulated silencer.
The REST binding sites derived from the human
synaptophysin and synapsin I gene are
functionally indistinguisable
To compare the biological activity of the NRSEs derived
from the synapsin I promoter and the first intron of the
human synaptophysin gene, we used model promoters
containing the luciferase gene as reporter, the SV40
promoter, and two copies of the NRSE derived from the
synapsin I and synaptophysin gene, respectively (reporter
Fig. 2. Activity of the human synapsin I and synaptophysin promoter in
neuronal and non-neuronal cells. (A) Schematic representation of the
synapsin I promoter/luciferase and synaptophysin promoter/luciferase
reporter genes pSyI
-2309/+47
luc and pSyp
-2356/+27
luc. (B) One of the
reporter plasmids pSyI
-2309/+47
luc or pSyp
-2356/+27
luc, was transfected
into NS20Y, SN56 (1 lg per plate) or 293T cells (0.5 lg per plate)
together with the reference plasmid pRSVb(NS20Y, SN56 cells:
0.5 lg per plate; 293T cells: 0.25 lg per plate) that encoded b-galac-
tosidase under control of the Rous sarcoma virus long-terminal repeat.
Forty-eight hours post-transfection cell extracts were prepared and the
b-galactosidase and luciferase activities of these extracts determined.
The data are presented as the ratio of luciferase activity (light units) to
b-galactosidase units (Aunits) measured in the cell extracts. At least
two experiments in quadruplicate were performed and the mean
±SEM is depicted.
Fig. 3. Localization of an NRSE in the human synaptophysin gene. (A)
Schematic representation of part of the human synaptophysin gene
containing the promoter region, exons I to III and introns I and II. The
location of the NRSE within the first intron of the gene is indicated.
The accession number for the human synaptophysin gene is U93305.
The NRSE encompassed nucleotides 29190–29210. (B) Sequence of
the NRSEs derived from the human synapsin I gene and the human
and rat synaptophysin genes.
FEBS 2003 Regulation of synaptophysin gene transcription (Eur. J. Biochem. 270)5
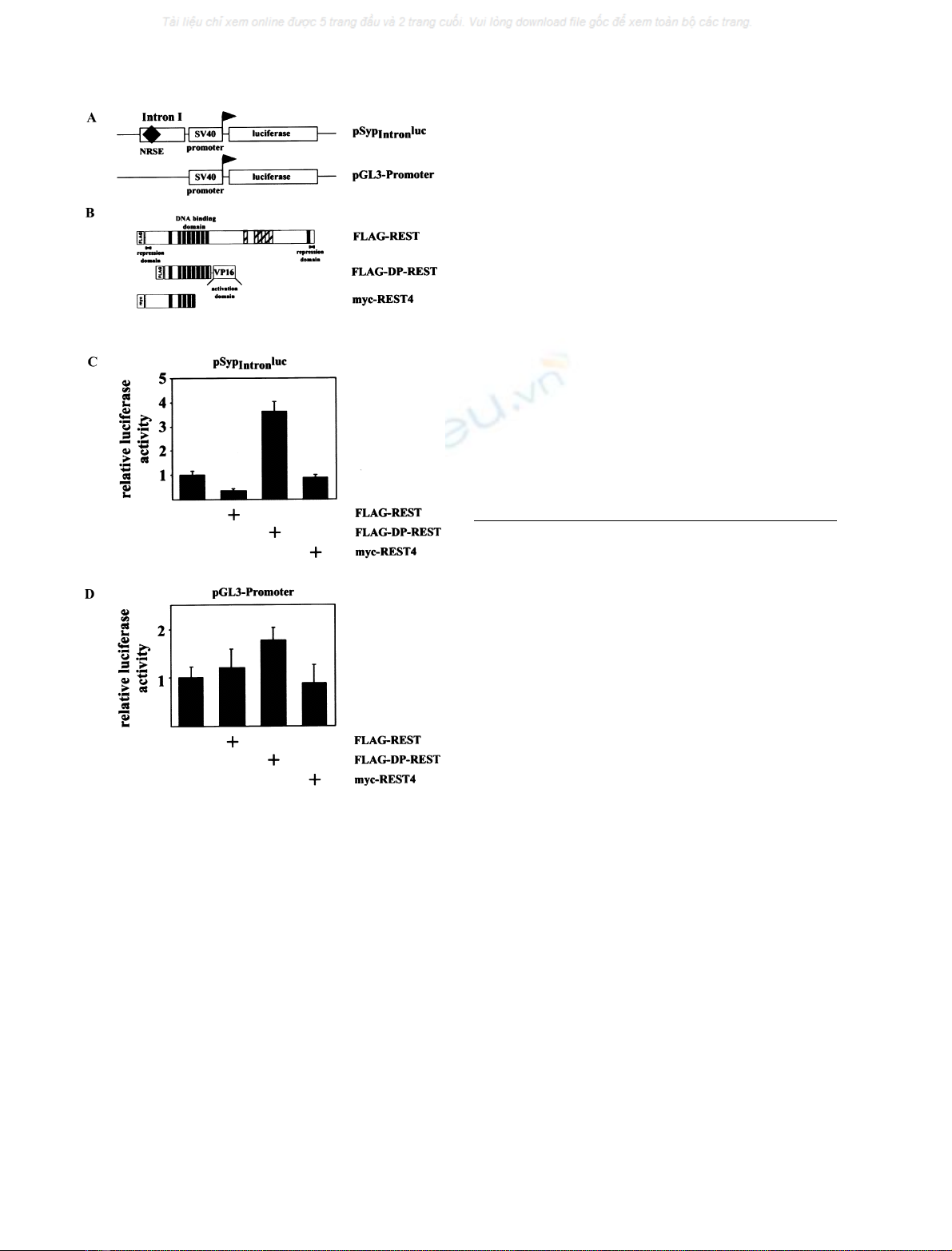
plasmids pSyINRSE
2
SV40luc and pSypNRSE
2
SV40luc,
Fig. 5A). One of the reporter plasmids, the internal
reference plasmid pRSVband expression vectors encoding
FLAG-REST, FLAG-DP-REST or myc-REST4 were
transfected into NS20Y neuroblastoma cells. Forty-eight
hours post-transfection, cells were harvested, cell extracts
prepared and the relative luciferase activities determined.
The results show that the presence of a REST binding site,
either from the synapsin I or synaptophysin gene, together
with expression of FLAG-REST, caused a striking decrease
in transcription (Fig. 5B, upper panels). Likewise, expres-
sion of FLAG-DP-REST increased reporter gene transcrip-
tion significantly (Fig. 5B, middle panels). The splice variant
of REST, REST4, however, did not activate or repress
transcription of NRSE-containing reporter genes (Fig. 5B,
lower panels), confirming previous results [16]. Taken
together, no major differences were detected between the
NRSE derived from the synapsin I or synaptophysin gene,
indicating that the NRSE functions in both genes as a
neuron-restrictive silencer element.
The zinc finger transcription factor Sp1 is responsible
for constitutive transcription via the human
synaptophysin promoter
The presence of a functional NRSE in the first intron of the
human synaptophysin gene indicates that REST blocks
human synaptophysin gene transcription through this
intronic neuron-specific silencer element. In contrast, no
neuron-specific genetic elements were found in the synapto-
physin 5¢-flanking region. Rather, this part of the transcrip-
tion unit contained constitutive transcriptional elements
that are active in neuronal as well as in non-neuronal cells.
The 5¢-flanking region of the human synaptophysin gene is
GC-rich, and potential binding sites for the transcription
factor Sp1 have been proposed [31]. To test whether Sp1 is
responsible for the constitutive transcriptional activity of the
human synaptophysin promoter, we used a dominant-
negative Sp1 mutant [17], consisting of the GST fused to the
DNA-binding domain of Sp1. Both domains were separ-
ated by a nuclear localization sequence, to ensure nuclear
targeting. As a control, an expression vector encoding a
nuclear-targeted GST was used (Fig. 6A). The expression
vectors were first transiently transfected into 293T cells.
The recombinant proteins were purified by glutathione
affinity chromatography and separated by SDS/PAGE.
Both proteins migrated on SDS/PAGE as expected.
Next, 293T and NS20Y cells were transfected with the
human synaptophysin promoter/luciferase reporter plasmid
pSyp
-2356/+27
luc, the GST-N encoding expression vector as
control, or increasing amounts of the expression vector
encoding GST-Sp1. The results show that the dominant-
negative Sp1 mutant decreased the constitutive transcrip-
tional activity of the synaptophysin promoter (Fig. 6C),
indicating that Sp1 is, at least in part, responsible for the
constitutive, tissue-unspecific activity of the 5¢-flanking
region of the human synaptophysin gene.
Fig. 4. REST regulates the transcription activity of a strong viral pro-
moter via the intronic NRSE derived from the human synaptophysin gene.
(A) Reporter plasmid pSyp
Intron
luc and pGL3-Promoter containing the
luciferase reporter gene and the SV40 promoter. Plasmid pSyp
Intron
luc
contains a fragment of the first intron of the human synaptophysin
gene, including the NRSE, 5¢of the SV40 promoter. (B) Modular
structure of FLAG-REST, FLAG-DP-REST, and myc-REST4. REST
contains a cluster of eight zinc fingers that function as DNA-binding
domain, and two repressor domains on the N- and C-termini of the
molecule. FLAG-DP-REST, a positive-dominant mutant of REST,
retains the DNA-binding domain, but lacks the repression domains and
has instead a transcriptional activation domain derived from the VP16
protein of herpex simplex virus. REST4 is a neuron-specific splice
variant of REST that contains the N-terminal repression domain and
five of the eight zinc finger motifs of the DNA-binding domain. In
addition, FLAG-REST, FLAG-DP-REST, and myc-REST4 contain
recognition sequences (triple FLAG tag or myc-tag) on the N-termini.
(C) One of the reporter plasmids pSyp
Intron
luc or pGL3-Promoter (1 lg
per plate), 0.5 lg per plate of the pRSVbinternal standard plasmid and
either 100 ng per plate of the emptyexpression vector pCMV5 or one
of the expression vectors encoding FLAG-REST, FLAG-DP-REST,
or myc-REST4 were introduced into NS20Y cells. Transcription was
analyzed by determination of the b-galactosidase and luciferase activ-
ities of the cell extracts.
6 M. Lietz et al. (Eur. J. Biochem. 270)FEBS 2003


























