
Identification and characterization of the
Escherichia coli
stress
protein UP12, a putative
in vivo
substrate of GroEL
Elena S. Bochkareva, Alexander S. Girshovich and Eitan Bibi
Department of Biological Chemistry, Weizmann Institute of Science, Rehovot, Israel
Many groups of proteins play important roles in the cell’s
response to various stresses. The molecular chaperone
GroEL of Escherichia coli represents one such highly con-
served family of stress proteins. We have observed that iso-
lated GroEL complexes from stationary cultures contain
various polypeptides that can be released from the chap-
eronin by GroES and/or ATP, and identified two such
polypeptides as the proteins GatY and UP12. Whereas
GatY had been isolated previously, as an in vivo substrate of
GroEL, the isolation of UP12 in a complex with GroEL was
intriguing, because based on sequence similarity it was sug-
gested that UP12 might also be a functional stress protein.
UP12 belongs to a family of universal stress proteins
(UspA family), of which UspA itself, and three additional
paralogues, have been characterized previously. Here we
show that UP12 accumulates under various growth inhibi-
tory conditions and induced by heat shock. Furthermore,
unlike wild-type cells, a UP12 deletion mutant recovers
slowly from late stationary growth conditions, and has a
marked sensitivity to the toxic agent carbonyl cyanide
m-chlorophenyl hydrazone (CCCP). Finally, coimmuno-
precipitation experiments confirmed the initial observation
that UP12 interacts with GroEL. Therefore, we suggest that
UP12 may function as a universal stress protein, interaction
of which with GroEL possibly ensures its proper folding
state.
Keywords: GroEL substrate; UP12; universal stress protein;
Stress response; E. coli.
Escherichia coli cells undergo a transition from a rapid
growth phase to a stationary phase, which is accompanied
by a variety of physiological changes that affect gene
expression, the structure and composition of the cell wall,
DNA organization, synthesis of storage compounds such as
glycogen and polyphosphate, and other cellular processes
[1,2]. As a result of these changes, the cells become resistant
to various deleterious stresses such as heat shock, UV
irradiation, acidic or basic conditions, osmotic shock, and
oxidation [3–5].
Studies carried out in several laboratories have identified
specific cellular networks and individual genes expressed in
the stationary growth phase that improve the survival of
E. coli during prolonged periods of starvation and other
stress conditions [6–11]. One of these genes is uspA,which
encodes a small cytoplasmic protein, UspA (universal stress
protein A) that is unique in its universal responsiveness to
diverse stresses [12]. The synthesis of UspA is greatly
increased under growth inhibitory conditions, including the
depletion of essential nutrients or exposure to various toxic
agents. Moreover, E. coli carrying an inactivated uspA is
more sensitive to prolonged growth inhibition caused by a
variety of starvation and other stress conditions [13,14].
In the course of systematically analyzing the sequenced
E. coli genome [15], it has been found that five ORFs share
some homologies with UspA. Two of them, encoded by
ybdQ and ynaF, were previously identified as unknown
proteins (UP12 and UP03, respectively) by 2D-PAGE [16].
Three E. coli paralogues of UspA have been characterized
recently [17], and the results of this study showed that UspA
is a prototype for a family of conserved proteins (universal
stress proteins) found not only in bacteria but also in other
organisms.
Other groups of proteins also play important roles in
bacterial stress response. One important group includes the
heat-shock proteins, whose induction under stress conditions
in E. coli requires the heat-shock transcription factor r
32
(rpoH gene product) [18]. Many heat-shock proteins, such as
members of the Hsp70 and Hsp60 protein families, are
molecular chaperones. Functionally, they bind to non-native
structural forms of various polypeptides and assist them in
reaching a native conformation [19]. Consequently, as
molecular chaperones, they prevent misfolding and aggre-
gation of unfolded proteins under heat-shock and other
stress conditions [20,21]. The E. coli heat-shock protein
GroEL belongs to the highly conserved Hsp60 family of
oligomeric molecular chaperones named chaperonins [22].
GroEL and its small cohort GroES were found to be essential
not only under stress, but also for growth under all
experimental conditions tested to date [23]. GroEL transi-
ently interacts (in a GroES- and MgATP-dependent manner)
with many unfolded newly synthesized proteins in vitro and
in vivo [24–26]. Among the proposed physiological substrates
of GroEL are structurally unstable proteins that require
GroEL for permanent conformational maintenance [27].
In the course of GroEL purification from stationary
cultures of E. coli, we noticed that a few polypeptides
Correspondence to E. Bochkareva, Department of Biological
Chemistry, Weizmann Institute of Science, Rehovot 76100, Israel.
Fax: + 972 89 344118, Tel.: + 972 89 342912,
E-mail: elena.bochkareva@weizmann.ac.il
Abbreviations: CCCP, carbonyl cyanide m-chlorophenyl hydrazone;
DNP, a-dinitrophenol; DM, n-dodecyl-b,
D
-maltoside; IPTG, isopro-
pyl b-
D
-thiogalactopyranoside.
(Received 15 February 2002, revised 25 April 2002,
accepted 3 May 2002)
Eur. J. Biochem. 269, 3032–3040 (2002) FEBS 2002 doi:10.1046/j.1432-1033.2002.02978.x
consistently co-sedimented with GroEL during sucrose
gradient ultracentrifugation. After incubation with GroES
and/or ATP, these polypeptides were released from the
chaperonin. One such protein that co-purified with GroEL
was identified as UP12. Based on limited sequence similarity
with UspA, we examined the possibility that UP12 itself
might be a stress protein. Here, we show that UP12 interacts
specifically with GroEL, and the results suggest that it plays
a role in the bacterial stress response. The possible
physiological relevance of UP12¢s interaction with GroEL
is discussed.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
The E. coli strains used in this work are MC4100
[F
–
araD139D(argF-lac)U169 rpsL150 relA1 flb5301 deoC1
ptsF25 rbsR], TG2 [F¢traD36 lacI
q
D(lacZ)M15 supE hsd D5
thi D(lac )proAB)D(srl-recA)306::Tn10 (tet
r)
], BL21(DE3)
[28] and BW25113 [29]. Cultures were grown aerobically in
liquid Luria–Bertani medium with ampicillin, 50 lgÆmL
)1
or kanamycin, 30 lgÆmL
)1
, when necessary. For carbon
starvation, cells were grown in M9 minimal medium with-
out amino acids [30], supplemented by a very low glucose
concentration (0.02%) compared with the usual concentra-
tion of 0.4%. For phosphate starvation, cells were grown in
TrisG medium [31] without amino acids supplementation
and with a limited concentration of KH
2
PO
4
(0.06 m
M
). As
a control, cells were grown with the normal concentration of
1.32 m
M
KH
2
PO
4
. Other growth conditions are described
below.
Isolation of GroEL and GroEL complexes
from cell extracts
E. coli TG2 bearing plasmid pOA encoding GroEL and
GroES [32] were grown at 37 C for 18–20 h in rich 2TY
medium containing ampicillin. Typically, 0.5 L of culture
was harvested and the pellet was resuspended in 7 mL of
buffer containing 30 m
M
Tris/HCI (pH 7.5), 60 m
M
KCI,
10 m
M
MgCI
2
,0.2m
M
EDTA, 1 m
M
dithiothreitol, and
0.5 m
M
phenylmethanesulfonyl fluoride. Cells were then
disrupted by treatment with lysozyme (1 mgÆmL
)1
)onice
for 10 min followed by a single rapid cycle of freeze and
thaw at 4 C. The entire cell lysate was then subjected to a
preparative sucrose gradient centrifugation as follows. Each
quarter of the lysate was loaded on top of a 36-mL 10–25%
sucrose gradient prepared in buffer A [40 m
M
triethanol-
amine-acetate (pH 7.5), 80 m
M
NH
4
Cl, 20 m
M
KCl, 10 m
M
MgCl
2
,0.1m
M
EDTA and 1 m
M
dithiothreitol]. After 21 h
centrifugation at 4 C (Beckman L8 ultracentrifuge, SW 27
rotor, 104 000 g) the middle fractions containing GroEL
were pooled and precipitated with ammonium sulfate. The
pellet was solubilized in buffer A and subjected to an
additional round of a preparative sucrose gradient centrif-
ugation in buffer B (same as buffer A, but with 450 m
M
NH
4
Cl instead of 80 m
M
). The GroEL-containing fractions
were pooled, diluted four times in buffer A and concentra-
ted using centriprep 30 (Amicon). The final GroEL
concentration was approximately 10 mgÆmL
)1
; aliquots
were frozen and stored at )80 C. Analytical ultracentrif-
ugations were carried out using 1.4 mL of a linear 5–20%
sucrose gradient in buffer A for 140 min at 4 C (Beckman
TL100 centrifuge, TLS55 rotor, 250 000 g). GroES was
purified as described previously [33].
The dissociation of polypeptides from their complex with
GroEL was tested by incubation (30 min at 25 C) of the
GroEL preparation in buffer A containing 8 m
M
ATP, with
or without GroES (GroES/GroEL ¼0.2 : 1, w/w) or 0.1%
n-dodecyl-b-
D
-maltoside (DM), followed by centrifugation
through a 5–20% sucrose gradient, as described above. The
top fractions containing free polypeptides were collected,
concentrated (using Centricon 30, Amicon) or precipitated
by 10% trichloroacetic acid. Samples were subjected to
SDS/PAGE and electroblotting on poly(vinylidene difluo-
ride) membranes (Bio-Rad). The membranes were then
stained for 0.5–1.0 min in 0.04% Coomassie R250 (pre-
pared in a solution of 50% methanol and 10% acetic acid).
Destaining was carried out in 50% methanol for 5–10 min
followed by an extensive wash with water. The appropriate
bands that corresponded to polypeptides X and Y were
excised and subjected to microsequencing analysis (Applied
Biosystems Procise Sequencer).
Immunoprecipitation of GroEL complexes from cell
extracts was carried out at 4 C for 3 h in buffer A,
containing 0.01% DM, using protein A–Sepharose pre-
loaded with affinity-purified anti-GroEL Ig. Blocking of
unspecific binding sites was accomplished by incubation
with BSA. After extensive washing with the same buffer,
proteins were eluted from the resin with buffer C [0.1
M
Tris/
HCI (pH 8.0), 1% SDS, 2 m
M
EDTA and 20 m
M
dithiothreitol] and analyzed by SDS/PAGE and Western
blotting.
Subcloning and deletion of the chromosomal
ybdQ
gene (encoding UP12)
The chromosomal ybdQ gene encoding UP12 (locus
AE000166, accession no. U00096 [15]), was amplified by
PCRusinga5¢complementary deoxyoligonucleotide
(5¢-CGCGGATCCATGTATAAGACAATCATTATGC-3¢)
containing a BamHI site (underlined nucleotides) and a 3¢
deoxyoligonucleotide harboring a HindIII site (5¢-CCCAA
GCTTTTAACGCACAACCAGCACC-3¢) as primers with
E. coli genomic DNA as a template and Taq polymerase
(Roche Molecular Biochemicals). Next, the purified PCR
product was ligated with plasmid pGEM-T Easy Vector
(Promega). E. coli HB101 cells were transformed with the
ligation mixture and a plasmid containing the ybdQ gene
insert was isolated. The BamHI–HindIII ybdQ fragment
was then isolated and subcloned into plasmid pET28a
(Novagen), which was digested with the same enzymes. The
resulting plasmid (pET28yQ) was isolated from E. coli
HB101 and the identity of the ybdQ insert was confirmed by
DNA sequencing. Finally, E. coli BL21(DE3)pLysS was
transformed with pET28yQ to overexpress UP12 as a
hybrid with an N-terminal extension containing a His
6
tag
separated from UP12 by a thrombin recognition site and
two flanking unrelated short sequences. The DNA sequen-
cing and the deoxyoligonucleotide synthesis were performed
by the Scientific Services Department of the Weizmann
Institute of Science.
Construction of ybdQ deletion E. coli mutant was carried
out as described previously [29]. A PCR product was
generated by using two 60-mer primers comprised of
FEBS 2002 UP12 is an E. coli universal stress protein (Eur. J. Biochem. 269) 3033
40 nucleotides homologous to regions adjacent to the
beginning and the end of ybdQ and additional 20-nucleotide
complementary to the template plasmid pKD13 carrying
the kanamycin resistance (kan) gene. The resulted linear
1.4-kb PCR fragment was gel-purified and introduced by
electroporation into E. coli cells BW25113 containing the
helper plasmid pKD46. Transformants were incubated 1 h
at 37 C and overnight at room temperature in SOC
medium and then plated on Luria–Bertani agar plates
containing kanamycin. Kanamycin resistant (Km
R
)trans-
formants were selected and colony-purified on agar plates
incubated overnight at 37 C. Mutant and wild-type cells
from single colonies were grown overnight in Luria–Bertani
broth at 37 C without an antibiotic and tested for loss of
the helper plasmid pKD46. The correct chromosomal
structure of DybdQ::kan mutant was verified by three
independent PCR experiments which were carried out with
two 20-nucleotide primers complementary to regions flank-
ing the ybdQ gene as a pair and in combination with two
kan– specific primers (k2 and kt; [29]). An additional PCR
experiment with the two primers used for subcloning of
ybdQ (see above), did not yield any product, as expected.
Finally, UP12 expression in the mutant and wild-type cells
was examined by Western blotting, as described below.
Purification of UP12 and preparation of anti-UP12 Ig
The UP12 hybrid containing a His
6
tag (see above) was
purified from E. coli BL21(DE3)pLysS(pET28yQ). Briefly,
cultures were grown in 2TY medium at 37 C and induced
with isopropyl thio-b-
D
-galactoside (IPTG; 1 m
M
)atthe
exponential growth phase (D
600
¼0.4) for 2.5 h. After
centrifugation, the cells were resuspended in 7.5 mL of
buffer K [20 m
M
Tris/HCI (pH 8.0) and 0.5
M
NaCl] and
disrupted by sonication using Microson (Heat Systems,
Inc.). The tagged UP12 was purified from the cell extract by
affinity chromatography using His-bind resin (Novagen),
according to the manufacturer’s instructions. The protein
was eluted with buffer K containing 1
M
imidazole and
cleaved by thrombin (Novagen) at room temperature for
17–20 h, using a protease/protein ratio of 1 : 800. The
protein concentration was measured using a Bradford
solution (Bio-Rad) and BSA as a standard. Polyclonal anti-
UP12 Ig were produced in rabbits by the Scientific Services
Department of the Weizmann Institute by a single injection
of 150 lg of the purified protein, followed by two booster
shots of the same amount of protein at 2-week intervals.
Serum was collected and used for immunoblotting at a
1 : 5000 dilution.
SDS/PAGE and Western blotting
In order to estimate the intracellular concentration of UP12
under various conditions, we centrifuged culture samples
containing equal amounts of cells (corresponding to
D
600
¼0.8). The pellets were washed with 0.2 mL of 10%
sucrose prepared in 10 m
M
Tris/HCI (pH 8.0) and then
lysed by adding 70 lL of buffer C (see above) containing
0.1 m
M
dithiothreitol instead of 20 m
M
. After incubation at
room temperature for 10 min, the lysates were centrifuged
to remove the pelleted DNA, and the protein concentrations
in the supernatants were measured using a Bio-Rad DC
protein assay. The supernatants were diluted 1 : 2 by
solution D (solution C with 20% glycerol, 0.002% bromo-
phenol blue and 30 m
M
dithiothreitol) and then incubated
at 86 C for 8 min. Typically, cell extracts (5–10 lgof
protein) were separated by SDS/PAGE using the standard
Laemmli system [30] with 13% and 4% (w/w) acrylamide in
the separating and stacking gels, respectively. In order to
estimate the ratio between the amounts of UP12 and the
total amount of protein in the extracts, a series of samples
containing determined amounts of the purified UP12 were
separated by SDS/PAGE along with the cell extract
samples. Immunoblots were performed according to the
ECL Western blotting protocol (Amersham), and the
chemiluminescence was detected by exposure of the mem-
branes to films. Protein quantities were estimated by scan-
ning densitometry using a Bio-Rad Imaging Densitometer
(Model GS-690).
RESULTS
Isolation of GroEL-polypeptide complexes
from a stationary-phase culture
The chaperonin GroEL comprises 14 identical subunits of
57.3 kDa and has a unique molecular mass of 800 kDa
that can be separated from almost all other E. coli proteins
by sucrose gradient centrifugation [19,24]. In order to isolate
GroEL accompanied by cytoplasmic proteins from station-
ary cultures, we prepared cell extracts from 20-h cultures of
E. coli TG2(pOA). The extracts were subjected to three
successive steps of sucrose gradient centrifugation. We
noticed that a few polypeptides consistently cosedimented
with GroEL and were found exclusively in the GroEL-
containing fractions after a third round of sucrose gradient
centrifugation (Fig. 1A), suggesting that these polypeptides
might be in vivo substrates of GroEL. Therefore, we
analyzed the effect of ATP and GroES on their dissociation
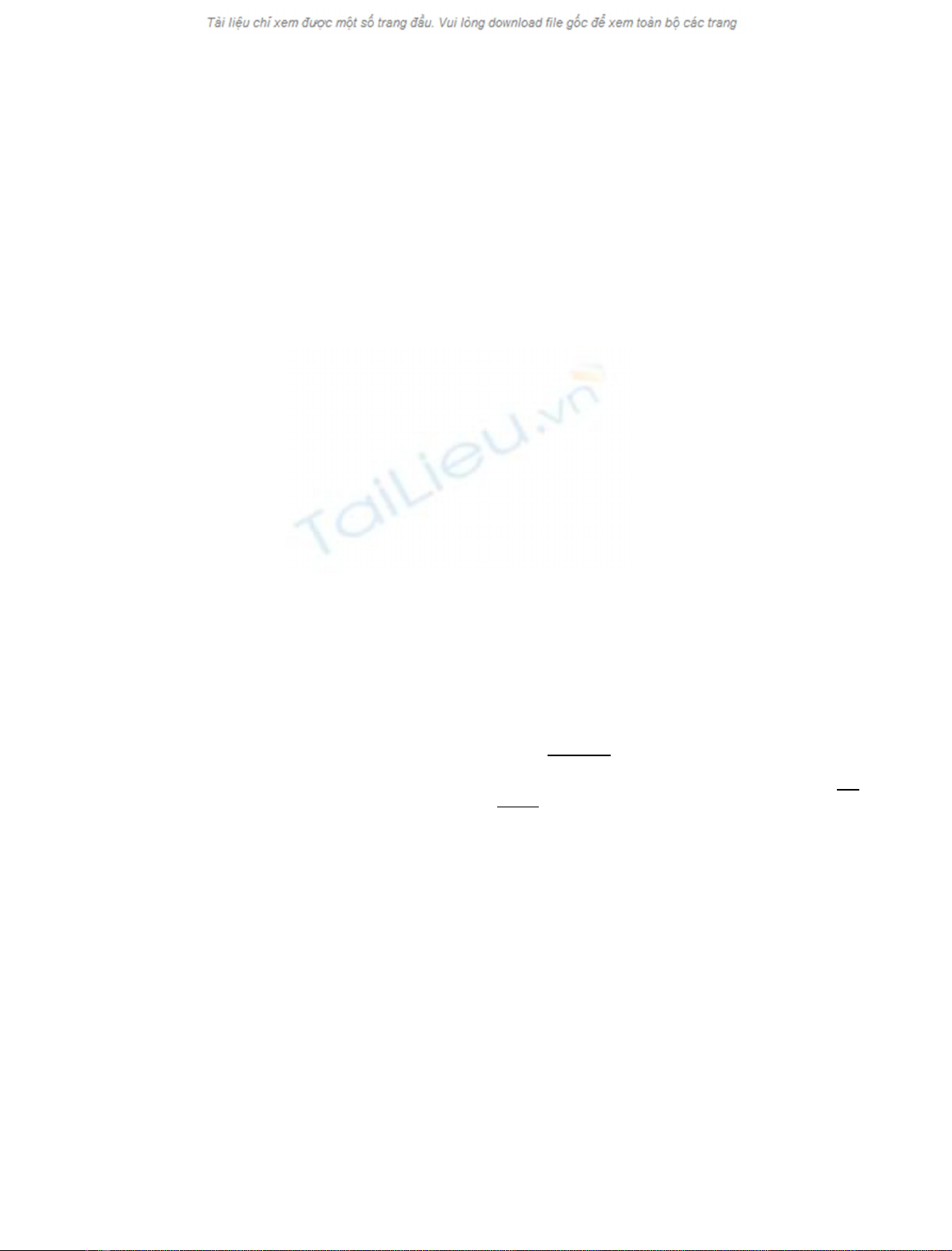
Fig. 1. ATP-dependent cosedimentation of proteins with chaperonin
GroEL. (A) Detection of polypeptides in the GroEL-containing frac-
tions of sucrose gradient. The crude preparation of GroEL (100 lg)
was subjected to analytical sucrose gradient centrifugation, as des-
cribed in Experimental procedures. Proteins in 14 fractions (100 lL
each) collected from the top of gradient were precipitated by TCA,
separated by SDS/PAGE and visualized by Coomassie staining. Lane
15 contains protein molecular mass markers. (B) The effect of ATP
preincubation on releasing of polypeptides from GroEL. Before
sucrose gradient centrifugation the GroEL preparation (40 lg) was
treated with 8 m
M
ATP (lane 1) or 8 m
M
ATP in the presence of 0.1%
DM (lanes 2–4) for 30 min at 25 C. Fractions (300 lL) was collected,
proceeded as in (A) and only some of them are presented. Lanes 1 and
2, as fractions 1–3 in (A), contain proteins recovered from the top of
sucrose gradient; lane 3 is intermediate fraction; and lane 4 corres-
ponding to fractions 7–10 in (A) contains oligomeric GroEL.
3034 E. S. Bochkareva et al. (Eur. J. Biochem. 269)FEBS 2002

from GroEL. As shown in Fig. 1B, a 16-kDa polypeptide
(Y) dissociates from GroEL by treatment with ATP,
whereas other polypeptides, including a 30-kDa polypeptide
(X) require both ATP and the co-chaperonin GroES for
dissociation (data not shown). Interestingly, the nonionic
detergent DM mimicked the effect of GroES by releasing
almost all of the polypeptides from GroEL. After treatment
with ATP and 0.1% DM, these polypeptides were recovered
in the top three fractions of the sucrose gradient (Fig. 1B,
lane 2). The ATP/GroES-promoted dissociation of these
polypeptides from GroEL, as well as the mild conditions
under which the cell extracts were prepared (see Experi-
mental procedures) suggest that the isolated GroEL com-
plexes may represent true physiologically relevant
interactions in stationarily grown cells.
Identification of proteins X (GatY) and Y (UP12)
In order to identify polypeptides X and Y, we repeated the
experiment (Fig. 1B) on a preparative scale. Briefly, after
their dissociation from the GroEL complex, proteins were
separated by SDS/PAGE, electroblotted onto poly(vinylid-
ene difluoride) membranes, and subjected to N-terminal
sequencing. The amino-acid sequences of proteins X and Y
were identified in the Swiss-Prot databank as GatY and
UP12, respectively. GatY (
D
-tagatose-1,6-bis-phosphate
aldolase of class II), is a homotetrameric protein consisting
of 31-kDa subunits; it belongs to a family of lyases involved
in carbohydrate metabolism. GatY is highly thermolabile
and is degraded in vivo at temperatures above 30 C [34].
Our results are in agreement with those of a recent work in
which GatY has been identified by other means as an in vivo
substrate of GroEL [27]. Interestingly, GatY is also included
in a list of proteins that aggregate at 42 CinE. coli
containing a dnaK deletion mutation (DnaK is a Hsp70
chaperone) [35]. Collectively, our observations and those of
others suggest that folding of GatY might require the
assistance of molecular chaperones, which bind to its
temperature-induced flexible conformation.
The second protein that was isolated in a complex with
GroEL was the 16-kDa protein UP12 (also termed UspG
[17]). This protein is encoded by the ybdQ gene and belongs
to the UPF0022 (UspA) protein family [15]. E. coli contains
five small members of this family including UspA itself
(Fig. 2), and one larger protein consisting of two UspA
domains in tandem [17]. The small members of this family
are proteins of 142–144 amino-acids long; they are acidic
and presumably located in the cytoplasm like UspA.
Members of this family share a strikingly similar hydro-
pathy profile (data not shown), and UP12 shares 27%
identical and similar residues with UspA. Taken together,
although the sequence similarity between UP12 and UspA
is not very pronounced, the following observations support
the classification of UP12 as a member of the UspA family.
Sub-cloning, purification, and characterization of UP12
and its interaction with GroEL
In order to investigate the suggestion that UP12 is a
functional member of the universal stress protein family and
further characterize its interaction with GroEL, we cloned
the UP12 encoding gene (ybdQ)byPCR.YbdQ was inserted
into the expression vector pET-28a, under the control of the
T7promoter. A UP12 hybrid protein of 21 kDa containing
an N-terminal His
6
tag was overexpressed, purified by
nickel-affinity chromatography, and treated with thrombin
(Fig. 3A). The resulting cleaved UP12 hybrid, which
contains a 15-residue N-terminal extension (Fig. 3A, lane
6), was used to raise antibodies in rabbits. Western blotting
revealed that the anti-UP12 Ig recognizes the two hybrid
forms of the isolated protein (before and after cleavage with
thrombin; Fig. 3B, lane 1). In addition, Western blotting of
the total E. coli extracts demonstrated that the antibodies
selectively recognize a 16-kDa protein that corresponds to
UP12 (Fig. 3B, lane 2).
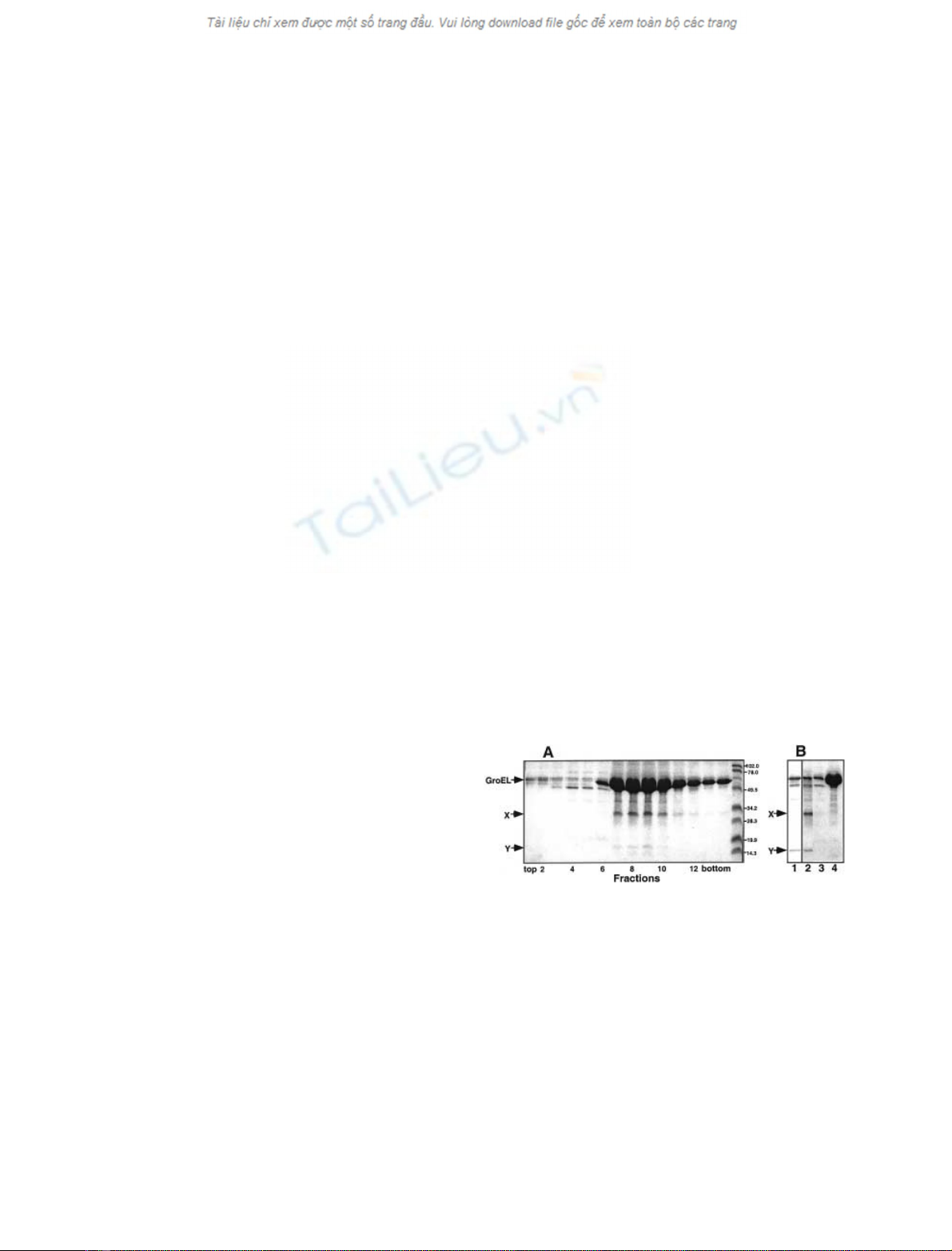
In order to investigate whether UP12 interacts with
GroEL in vivo, we analyzed GroEL complexes by co-
immunoprecipitation. GroEL complexes were isolated from
late stationary cultures of E. coli TG2(pOA) overexpressing
GroEL by immunoprecipitation with anti-GroEL Ig. As
showninFig.3C,35% of the UP12 were co-immuno-
precipitated with the GroEL. Remarkably, upon treatment
of the extracts with ATP, UP12 is completely released from
the GroEL complex (Fig. 3C, lanes 2 and 4). When extracts
prepared from late stationary cultures of E. coli MC4100 or
TG2 that do not overexpress GroEL were subjected to a
similar analysis, 4–5% of UP12 was found in a complex
with GroEL (data not shown). Similar yields were obtained
previously for some of in vivo GroEL substrates isolated
from the exponentially grown cells by immunoprecipitation
with anti-GroEL Ig [27]. The high yield of the UP12-GroEL
complex isolation and the ATP-mediated dissociation of
the complex strongly support the suggestion that the two
stress proteins GroEL and UP12 interact with each other
in vivo, and that this interaction might be physiologically
relevant.
Fig. 2. Sequence alignment of members of the E. coli UspA family. Optimal alignment of the best regions of similarity among the sequences was
performed using the program
PRETTYBOX
(Wisconsin
GCG
package, Version 10). A black or a gray background indicates identical and similar
residues, respectively. Swiss-Prot accession numbers are as follows: UspA, P28242 [12], YiiT, P32163; UP03, P37903; UP12, P39177; YecG, P46888
[15].
FEBS 2002 UP12 is an E. coli universal stress protein (Eur. J. Biochem. 269) 3035
UP12 is highly expressed at the stationary phase and
under conditions of phosphate or carbon starvation
To explore the properties of UP12 as a possible stress
protein, we analyzed the level of UP12 expression in E. coli
cells cultured in various media and under different experi-
mental conditions. Cultures of E. coli MC4100 were grown
at 37 C in Luria–Bertani, and samples were withdrawn at
the indicated times (Fig. 4A). Cell extracts were then
subjected to SDS/PAGE and electroblotting, and the
relative amount of UP12 in each sample was estimated by
semiquantitative Western blotting (Fig. 4B). As shown,
UP12 is hardly expressed during exponential growth; it
starts to accumulate at the early stationary phase, and its
steady-state level increases further during the late stationary
phase. As a result of this accumulation, the relative amount
of UP12 in stationary cells is about 10 times higher than that
observed at the beginning of growth (Fig. 4A). For
comparison, the amount of GroEL was determined in the
same extracts using anti-GroEL Ig. It has been previously
shown that at the beginning of the stationary phase the rate
of synthesis of heat-shock proteins increases considerably,
but only transiently [2,18]. Similarly, we observed that the
steady-state amount of GroEL remains constant during
growth, with only a slight increase (twofold) at the
stationary phase (Fig. 4B).
As shown, UP12 accumulates in cells grown in Luria–
Bertani during the stationary growth phase, possibly due to
nutrient exhaustion. In order to examine whether UP12 is
induced by starvation, we tested the expression of UP12 in
cells grown in minimal media containing limited concen-
trations of phosphate or a carbon source. The amount of
UP12 increased dramatically under both starvation condi-
tions (Fig. 4C). With phosphate starvation, UP12 accumu-
lation follows the arrest of growth, whereas under carbon
starvation conditions an 1 h delay is observed (Fig. 4C).
Interestingly, in supplemented minimal media, unlike in
Luria–Bertani, UP12 accumulation occurs only after pro-
longed (10–15 h) incubation of growth ceasing cells
(Fig. 4C and data not shown). Taken together, our results
indicate that the accumulation of UP12 is not due to a
certain stress caused by exhausting a specific nutrient in the
medium, but rather as a result of general growth inhibitory
conditions.
UP12 expression is induced in response to toxic agents
and increased temperature
Next, we examined the effect of toxic agents on UP12
expression. As shown in Fig. 5, the addition of DNP or
CCCP to exponential cultures led to an immediate arrest of
growth followed by an increased expression of UP12. In the
presence of DNP, the induction of UP12 was somewhat
slower compared with the rapid response to CCCP. In both
cases, however, after prolonged incubation with the toxic
compounds (4–5 h), the steady-state amount of UP12
increased up to fivefold its amount in untreated cells
(Fig. 5A,B). In order to test the expression of UP12 under
cold or heat shock conditions, cultures were grown at 37 C
in Luria–Bertani, and then transferred at the mid-log phase
to either 30 Cor44C.AsshowninFig.5C,therateby
which UP12 expression was increased at 30 C is similar to
that at 37 C. In contrast, heat shock at 44 C induced a
remarkably rapid accumulation of UP12. Therefore,
increased synthesis of UP12 occurs not only in growth-
arrested cells but also under heat shock conditions.
The
E. coli
DybdQ mutant shows a reduced growth rate
during stationary-phase-exit and an increased sensitivity
to CCCP
In order to study the possible biological function of UP12, an
E. coli mutant deleted of the UP12 encoding gene (ybdQ)was
explored. The mutated DybdQ::kan strain was constructed as
described previously [29] with the E. coli strain BW25113.
This strain behaves as E. coli MC4100 or TG2, with regard
to its UP12 expression pattern (data not shown), and as
Fig. 3. Purification of His
6
–UP12, characterization of the anti-UP12 Ig
and coimmunoprecipitation of UP12 with GroEL from cell extracts. (A)
Purification of UP12. His
6
–UP12 as a hybrid with an N-terminal
extension containing a His
6
tag separated from UP12 by a thrombin
recognition site and two unrelated short sequences was purified from
E. coli BL21(DE3)pLysS cells harboring pET28yQ by affinity chro-
matography on His-bind resin, as described in Experimental proce-
dures. Fractions of 15 mL of washing solution and 2.5 mL of eluates
were collected and 0.15% of each fraction was subjected to SDS/
PAGE. After electrophoresis the gel was stained with Coomassie Blue.
Lane 1, total cell extract; lanes 2, 3 and 4, column wash fractions (with
5m
M
,60m
M
and 90 m
M
imidazole, respectively); lane 5, elution
fraction (with 1
M
imidazole); lane 6, purified His
6
-UP12 after cleavage
by thrombin. Lane 7 contains protein markers. Arrow indicates the
position of a hybrid protein, His
6
–UP12. (B) Western blotting with
anti-UP12 Ig. Lane 1, purified His
6
–UP12 (10 ng) after incomplete
cleavage with thrombin; lane 2, total cell extract (5 lg protein) pre-
pared from E. coli MC4100 grown in Luria–Bertani medium for 24 h
at 37 C. Arrow indicates the position of UP12. (C) Co-immunopre-
cipitation of UP12 with GroEL from cell extracts. Isolation of GroEL
complexes from the E. coli TG2(pOA) cell extracts using protein
A–Sepharose preloaded with affinity-purified anti-GroEL Ig were
performed as described in Experimental procedures. UP12 coimmu-
noprecipitatedwithGroELfrom15and30 lg of the cell lysate without
(lanes 1 and 3) or in the presence of ATP (lanes 2 and 4) was detected
with anti-UP12 Ig. Samples (3 and 6 lg of total proteins) of the cell
extract that was not immunoprecipitated are shown in lanes 5 and 6.
3036 E. S. Bochkareva et al. (Eur. J. Biochem. 269)FEBS 2002


























