
Characterization of a cathepsin L-associated protein in
Artemia
and its relationship to the FAS-I family of cell adhesion proteins
Alden H. Warner
1
, Ervin Pullumbi
1
, Reinout Amons
2
and Liqian Liu
1
1
Department of Biological Sciences, University of Windsor, Windsor, Ontario, Canada;
2
Department of Molecular Cell Biology,
Sylvius Laboratory, Leiden, the Netherlands
We reported previously that the major cysteine protease in
embryos and larvae of the brine shrimp, Artemia franciscana,
is a heterodimeric protein consisting of a catalytic subunit
(28.5 kDa) with a high degree of homology with cathep-
sin L, and a noncatalytic subunit (31.5 kDa) of unknown
function. In the study reported here the noncatalytic subunit,
or cathepsin L-associated protein (CLAP), was separated
from cathepsin L by chromatography on Mono S and
found to contain multiple isoforms with pIs ranging from 5.9
to 6.1. Heterodimeric and monomeric cathepsin L showed
similar activity between pH 5 and 6.5, while the heterodimer
was about twice as active as monomeric cathepsin L below
pH 5. The heterodimer was more stable than the monomer
between pH 6 and 7.4 and at 30–50 C. Artemia CLAP and
cathepsin L are present in nearly equimolar amounts at all
stages in the life cycle and most abundant in encysted eggs
andembyros.Moreover,CLAP,eitherfreeorasacomplex
with cathepsin L, was resistant to hydrolysis by cathepsin L.
Two clones coding for CLAP were isolated from an Artemia
embryo cDNA library and sequenced. Both clones have
nearly identical open reading frames, but show differences at
the 5¢-and3¢-termini. Each cDNA clone has an extensive
3¢-untranslated region containing 70–72% A+T. The
deduced amino acid sequence of CLAP cDNA revealed two
domains which were very similar to domains in fasciclin I
and other cell adhesion proteins. The nucleotide sequences of
clones 1 and 2 have been entered into the NCBI database
(AY307377 and AY462276). This study supports the view
that the noncatalytic subunit of the heterodimeric cysteine
protease in Artemia stabilizes cathepsin L at various pH and
temperatures normally inconsistent with cathepsin L from
other organisms, and that CLAP serves as a docking
mechanism for cathepsin L at nonlysosomal sites in Artemia
embryos.
Keywords:Artemia; cathepsin L; cell adhesion proteins;
fasciclins.
Cathepsin L (CL) is a ubiquitous cysteine protease in
eukaryotes and essential for development in several organ-
isms including Xenopus laevis [1], Caenorhabditis elegans [2],
and Artemia franciscana [3]. Inhibition of CL activity in
these organisms, or deletion of the CL gene, leads to severe
abnormalities and even death. Developmental events
dependent on cysteine protease activity are numerous and
include yolk utilization [3–5], activation of latent enzymes
[6], gastrulation [1], differentiation [7–9], tissue remodelling
[10], implantation [11], and molting [3,12,13]. In developing
embryos, cysteine proteases are often found in the cyto-
plasm and extracellular matrix where they may have
regulatory functions, unlike in somatic cells of multicellular
organisms where these enzymes are primarily lysosomal
and thought to play a role in intracellular protein turnover
and degradation [14,15]. In mammals, cysteine proteases
may function in transcription factor regulation [16], in
antigen processing [17], and in several parasitic organisms
cysteine proteases are considered to be virulence factors
because they are secreted at the site of invasion [18,19].
Over-expression and secretion of cysteine proteases is also
common in various pathological conditions [20–22].
In embryos and larvae of the brine shrimp, A. franciscana,
the major protease is a heterodimeric cathepsin L-like
protease (CLP) consisting of a catalytic subunit (CL) of
28.5 kDa and noncatalytic subunit of 31.5 kDa with a total
molecular mass of 60 kDa [23,24]. The catalytic subunit of
the complex has a high degree of homology with cathepsin L
from several sources [24]. The noncatalytic subunit (cathep-
sin L-associated protein; CLAP) has, in vitro, a high affinity
for monomeric CL, and together, they form a heterodimeric
protease which has been resolved into seven isoforms with pI
values ranging from 4.6 to 6.2 [24]. Both subunits of CLP are
glycosylated; the catalytic subunit contains O-linked carbo-
hydrates and the noncatalytic subunit contains N-linked
carbohydrate [24]. Cell fractionation and immunocyto-
chemical studies of Artemia embryos and larvae indicate
that about 85% of the protease is nonlysosomal with
considerable antibody stain appearing at the surface of yolk
platelets and in the extracellular matrix [3,25].
cDNAs encoding the CL subunit of Artemia CLP
have been isolated and sequenced and their amino acid
Correspondence to A. H. Warner, Department of Biological Sciences,
University of Windsor, Windsor, Ontario, N9B 3P4, Canada.
Fax: + 519 971 3609, E-mail: warner1@uwindsor.ca
Abbreviations: CL, cathepsin L, catalytic subunit, monomer; CLP,
cathepsin L-like protease, dimer; CLAP, cathepsin L-associated
protein; PI-PLC, phosphatidylinositol-specific phospholipase C;
GPI, glycosyl-phosphatidylinositol; CNBr, cyanogen bromide;
TNBS, trinitrobenzenesulfonic acid.
(Received 23 April 2004, revised 19 July 2004,
accepted 19 August 2004)
Eur. J. Biochem. 271, 4014–4025 (2004) FEBS 2004 doi:10.1111/j.1432-1033.2004.04338.x

composition deduced [24]. At the amino acid level, Artemia
CL has 73.9% identity with Drosophila CL and 68.7%
identity with human CL. Despite the high degree of
similarity with Drosophila, human and other cathepsin Ls,
Artemia CL appears to function as a heterodimer (i.e., CLP)
of 60 kDa and not as a monomeric protein like in other
eukaryotes. Until now the noncatalytic subunit of CLP (i.e.,
CLAP) has received little attention.
This report focuses mainly on characterization of CLAP
and its potential role in the function of CL. Herein, we
present evidence that CLAP enhances the stability of CL to
temperatures and pH normally inconsistent with CL
activity. Primary sequence analysis of CLAP and cDNA
clones coding for CLAP show it to be a cell adhesion
protein and member of the fasciclin I family of proteins.
These results support the hypothesis that CL in Artemia
embryos is unique and functions outside lysosomes, in the
cytoplasm and extracellular matrix, unlike CL in many
other higher eukaryotes.
Materials and methods
Purification of cathepsin L-like protease
The cathepsin L-like protease (CLP) in embryos of the
brine shrimp, A. franciscana was purified using a modifica-
tion of a published method [24]. Fifty grams of fully
hydrated Artemia cysts were homogenized in ice-cold
homogenization buffer (50 m
M
Tris/HCl, pH 7.2, 5 m
M
KCl, 1 m
M
dithiothreitol and 10 m
M
MgCl
2
)usinga
motorized mortar and pestle (Torsion Balance Co, Clinton,
NJ, USA). Following centrifugation to remove nuclei, yolk
platelets, mitochondria (10 000 g, 20 min) and ribosomes
(105 000 g, 2.5 h), the soluble material was treated with
solid ammonium sulfate to obtain the 35–75% ammonium
sulfate insoluble material. The latter was collected by
centrifugation, dissolved in Buffer A [15 m
M
potassium
phosphate, pH 6.8, 25 m
M
KCl and 10% (w/v) glycerol],
then desalted on a column of Sephadex G-25 using Buffer A
as the eluent. The protease was purified to near homo-
geneity by sequential chromatography on DEAE–Seph-
arose, Concanavalin A–Sepharose, Superose 12 and Mono
Q [23,24]. The major isoforms of Artemia CLP that eluted
from the Mono Q column were combined and concentrated
to about 1 mL using Centricon 10 filters (Amicon Canada,
Oakville, ON, Canada). All chromatographic media were
from Amersham Pharmacia Biotech (Baie d’Urfe, QC,
Canada).
Protein and protease assays
The protein content of all column fractions was determined
by the Bio-Rad microassay [26] or bicinchoninic acid assay
[27] using BSA as the protein standard. Cysteine protease
activity of column fractions was determined using protamine
sulfate as substrate and the trinitrobenzene sulfonic acid
(TNBS) method [23]. One unit of protease activity was
defined as the release of 1 micromole of amino peptide per
minute from the substrate at pH 4.0 and 40 C. CL assays
were carried out using a modified method of Barrett &
Kirschke [28].All reaction vessels contained the following:
0.2 m
M
Cbz-Phe-Arg-4-methoxy-b-naphthylamide, 83 m
M
potassium phosphate, pH 5.0, 0.67 m
M
EDTA, 0.5 m
M
dithiothreitol, and 35–100 pmol of enzyme. The reaction
also contained dimethylsulfate (1.0–1.5%) in which the
substrate was dissolved. At the desired incubation time an
aliquot of the reaction mixture was added to an equal volume
of coupling buffer [5 m
M
mersalyl acid, 30 m
M
NaOH, 2%
(v/v) Brij and 0.81 m
M
EDTA, adjusted to pH 4.0 with 1
M
HCl] to which was added an additional volume of coupling
buffer containing 0.5 mgÆmL
)1
Fast Garnet (Sigma, Mis-
sissauga, ON, Canada). After 15 min incubation at room
temperature, the complex was extracted with 1 mL n-butanol
and the color intensity determined by analysis at 520 nm.
The number of pmoles of cathepsin L were determined by
titration of the active site with E-64 as described previously
[29]. The concentration of heterodimeric cathepsin L was
64–65% of that calculated from the protein concentration,
while monomeric cathepsin L was 60–61% of the calculated
value based on protein content. Rate constants were
calculated as pmol b-naphthylamine released per minute
per pmol of active protease at pH 5.0 and temperature indi-
cated. Artemia p26 protein was a gift of T. MacRae
(Dalhousie University, Halifax, NS, Canada), while the
protein artemin was prepared from Artemia cysts [30].
Isoelectric focussing and sodium dodecylsulfate
polyacrylamide gel electrophoresis
Isoelectric focussing (IEF) was performed in glass tubes
(0.5 ·12 cm) containing 6% (w/v) acrylamide, 2% (v/v)
4/6 ampholytes (Bio-Lyte; Bio-Rad, Mississauga, ON,
Canada), 1% (v/v) 3/10 ampholytes (Bio-Lyte), and 12.5%
(v/v) glycerol using a Haake–Buchler unit (Baxter, McG-
raw Park, IL, USA). The protein samples contained 10%
(v/v) glycerol, 0.1% (v/v) 3/10 ampholyte, 0.002% (w/v)
bromphenol blue and either CLAP or IEF standards (pI
4.45–9.6) in a final volume of 0.1 mL. The top buffer
(catholyte) was 100 m
M
NaOH and the bottom buffer
(anolyte) was 3 m
M
indole-acetic acid. Isoelectric focussing
was initiated at 350 V and 1.5 mA per gel column, and the
focussing was completed by 18 h at 4 C. The ampholytes
and IEF standards were from Bio-Rad. Following electro-
phoresis, the gels were soaked in several changes of distilled
water for about 10 min then stained with the Bio-Rad
silver reagent as recommended by the supplier. A control
gel containing buffer in place of protein was washed briefly
in distilled water, then 0.5 cm sections were placed in
1.0 mL distilled water for pH measurement. Gels contain-
ing the IEF standards and buffer only gave identical linear
responses with gel length. In a separate experiment, CLAP
was treated with phosphatidylinositol-specific phospho-
lipase C (PI-PLC) (Sigma) prior to analysis by IEF to test
for glycosyl-phosphatidylinositol (GPI) units in the protein
[31].
SDS/PAGE was performed in 12% (w/v) acrylamide gels
[32]. Following electrophoresis, gels were stained for 1 h
with 0.1% (v/v) Coomassie blue R-250 in 40% (v/v)
methanol and 10% (v/v) acetic acid then destained
overnight in 5% (v/v) methanol and 7.5% (v/v) acetic acid.
Acrylamide gels containing various preparations of CLP
and its subunits were also stained with Pro-Q Diamond
phosphoprotein stain (Molecular Probes, Eugene, OR,
USA) according to the manufacturer’s instructions.
FEBS 2004 Cathepsin L and cell adhesion protein in Artemia (Eur. J. Biochem. 271) 4015
Cysteine protease analysis at different stages
in the
Artemia
life cycle
Harvested organisms were reared in the laboratory to the
desired stage in their life cycle [3,33]. At the desired stage,
intact organisms were washed with distilled water, blotted of
excess water then frozen by immersion in liquid nitrogen.
Ovisacs from adult females containing encysted embryos or
nonencysted embryos were removed with a scalpel while
frozen in liquid N
2
. Gravid females from which the ovisacs
had been removed were saved for analysis. Immature,
nongravid females containing no visible signs of eggs, and
adult males, were collected, washed and frozen in liquid N
2
.
All tissues were stored at )70 C until needed. The frozen
tissues were homogenized in a buffer containing 50 m
M
sodium phosphate, pH 7.4, 1 m
M
EDTA and 5% (w/v)
SDS (at 70 C) using small glass homogenizers. The
insoluble material was removed by centrifugation, and
aliquots were taken for protein measurement and analysis in
7–18% SDS/PAGE gels. The amounts of catalytic and
noncatalytic subunits of CLP in each tissue extract were
determined by densitometry as described previously [25].
Amino acid sequencing of CLAP and CLAP fragments
Mono S purified and untreated CLAP was subjected to
Edman sequencing on a Hewlett–Packard G1005A pro-
tein sequencer. A cyanogen bromide (CNBr) generated
peptide of CLAP of about 25 kDa was purified by SDS/
PAGE, transferred to a poly(vinylidene difluoride) mem-
brane and sequenced by Edman degradation along with
five peptides obtained by Lys-C treatment of CLAP
(Eastern Quebec Peptide Sequencing Facility, Ste-Foy,
QC, Canada). In addition, pool sequencing, i.e. sequen-
cing of the complete mixture of CNBr-generated peptides,
was also performed.
Isolation and sequencing of cDNA clones encoding CLAP
A cDNA library prepared from cysts of A. franciscana was
a gift from T. MacRae (Dalhousie University, Halifax, NS,
Canada) prepared originally by L. Sastre (Instituto de
Investigaciones Biome
´dicas, CSIC/UAM, Madrid, Spain).
The library was constructed in phage kZAP II (Stratagene,
La Jolla, CA, USA) with the cDNAs were inserted between
the EcoRI and XhoI sites in the multiple cloning region of
the vector. The phage were amplified in XL1-Blue-MRF¢
(Stratagene) then probed with a
32
P-labeled 564 bp PCR
product generated using primers constructed from amino
acid sequence data of CLAP, and cloned into pCR2.1
(Invitrogen, Burlington, ON, Canada). Approximately
2·10
6
plaques were screened using standard protocols
[34], and six plaques, identified by hybridization to the
564 bp PCR product, were chosen for further analysis.
The isolated phage were converted to Bluescript phage-
mids using ExAssist helper phage and a protocol provided
by the supplier (Stratagene). Six cDNA clones were grown
overnight in the presence of ampicillin (50 lgÆmL
)1
)andthe
DNA was isolated using the Wizard Miniprep Kit (Promega,
Madison, WI, USA). All clones showed identical restriction
maps, and two were sequenced by cycle sequencing using
primers constructed from the original 564 bp PCR product
and from information in the Bluescript phagemid. Sequen-
cing was performed on a Visible Genetics (Suwanee, GA,
USA) instrument using the Thermo Sequenase Cy5.5
Terminator Cycle Sequencing Kit (Amersham Pharmacia).
Results
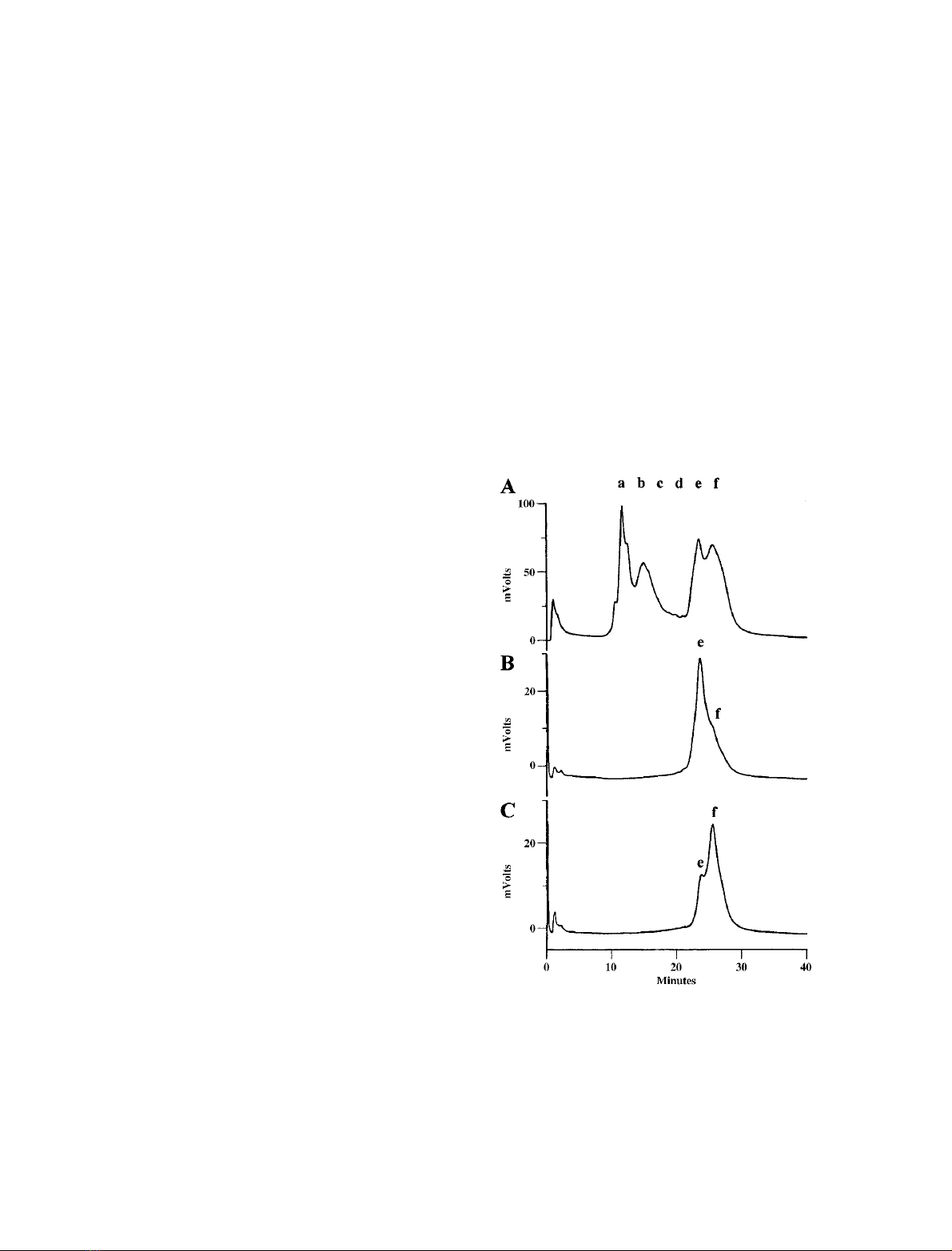
Separation of
Artemia
CLP subunits by HPLC on Mono S
Various fractionation methods have been attempted to
separate the catalytic and noncatalytic subunits of Artemia
CLP without loss of protease activity, but none has been
successful except cation-exchange chromatography. How-
ever, partial separation of Artemia CLP subunits was
achieved by chromatography on Mono S following pre-
incubation of the complex at pH 5 for at least 2.5 h at
0–4 C, including dialysis (Fig. 1A). This step resulted in
two, partially separated, fractions of CLAP (eand f)which
could not be resolved completely by re-chromatography on
MonoS(Fig.1B,C).
Fig. 1. Fractionation of Artemia cathepsin L subunits by HPLC on a
Mono S column. Prior to chromatography on Mono S 0.9 mg of
purified heterodimeric Artemia cathepsin L was adjusted to pH 5.0
with 1
M
sodium acetate, pH 4, incubated for 1 h at 0 C, then dia-
lyzed against Buffer A. (A) Elution profile of the Artemia cathepsin L
subunits monitored at 280 nm and expressed in mV on the yaxis.
Column fractions in the region of a–fwere concentrated for protease
assays and subunit analysis by SDS/PAGE (Fig. 2A,B). (B,C) Frac-
tions eand fwere re-chromatographed on the Mono S column under
conditions identical to those used initially.
4016 A. H. Warner et al.(Eur. J. Biochem. 271)FEBS 2004
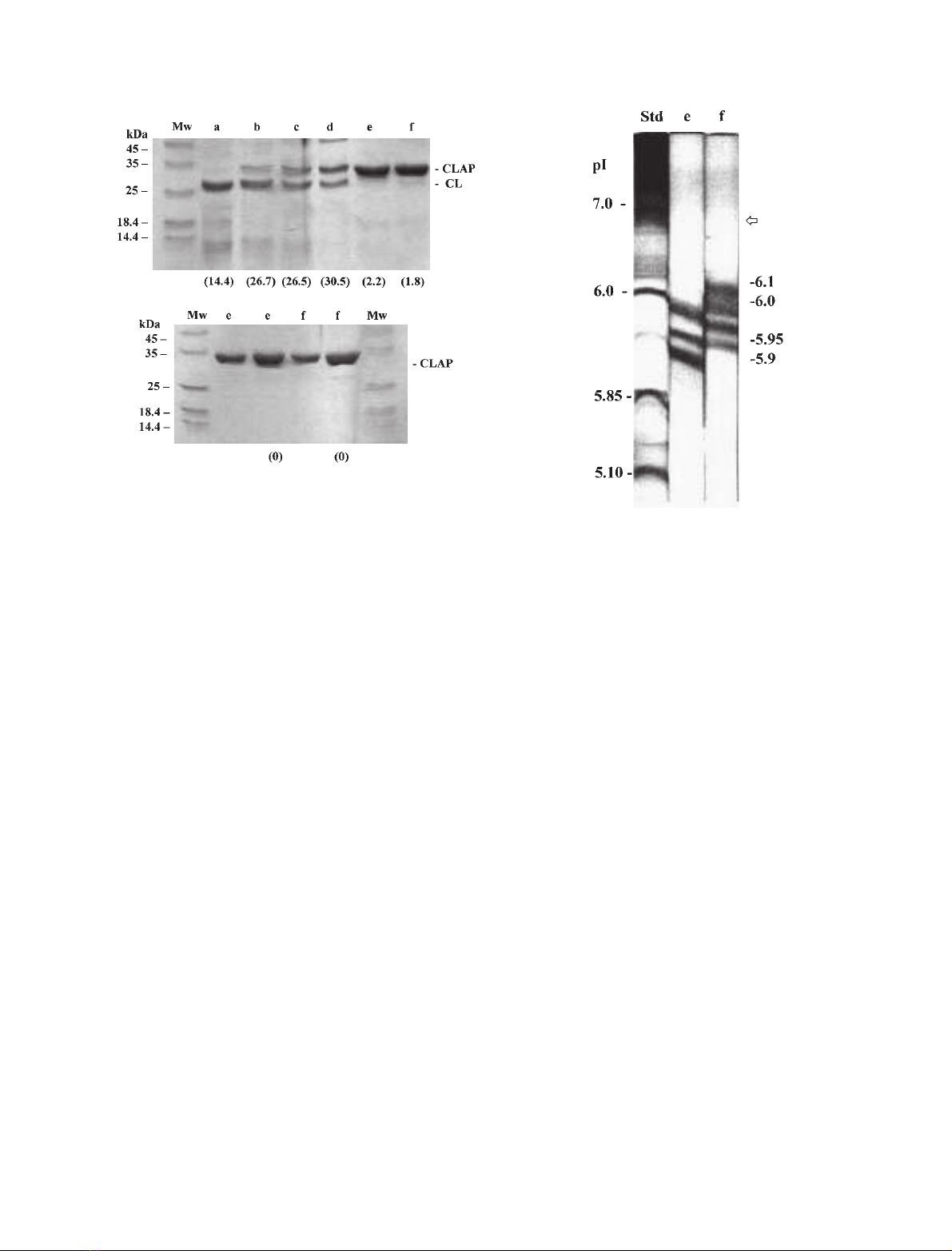
The protein composition of Mono S fractions a–fwere
analyzed by SDS/PAGE (Fig. 2). The main protein in
fraction awas the catalytic subunit of 28 kDa, while peaks
or areas labelled b, c and dcontained both subunits.
Column fractions b, c and dprobably represent specific
undissociated isoforms of the heterodimeric CLP, as each
contained both subunits of the native protease. Gel lanes e
and fcontained mainly CLAP of molecular mass 31.5 kDa.
The residual protease activity in peaks eand fdisappeared
during re-chromatography on Mono S (Fig. 2B). Treat-
ment of an SDS/PAGE gel containing CLP and CLAP with
a phosphoprotein stain did not reveal phosphate additions
to these proteins. Only lanes in the gel containing the known
phosphoproteins ovalbumin, b-casein and pepsin gave a
reaction. Thus, while Artemia CLAP fractions eand fare
clearly distinguishable on Mono S, they have identical
molecular masses (31.5 kDa), and they are devoid of
phosphate linked to Ser, Thr and Tyr.
Analysis of
Artemia
CLAP by isoelectric focussing
CLAP fractions eand f(Fig. 1B,C) were analyzed by IEF.
Fractions eand fshowed three and four bands, respectively,
on IEF gels with pI values ranging from 5.9 to 6.1 (Fig. 3).
Fractions eand fhave at least one unique isoform each (pI
5.9 for eand pI 6.1 for f), while two bands of pI 5.95 and pI
6.0werecommontoeachofthemajorCLAPfractions,
although this does not mean that these are identical
isoforms. Overall, Artemia CLAP appears to contain four
isoforms in nearly equal amounts, but these isoforms were
not resolved by chromatography on a C-18 reverse phase
column in which fractions eand fshowed identical elution
characteristics using acetonitrile/trifluoroacetic acid as the
eluent (data not shown and [24]).
Activity of dimer and monomer forms of
Artemia
CLP
at different pH and temperatures
Freshly prepared Artemia CLP (60 kDa, dimer) and CL
(28.5 kDa, monomer) (Fig. 1A, peak a) were assayed for
CL activity in parallel reaction vessels at 30 C and various
pH (Fig. 4A). The monomer showed maximum activity at
pH 5.0, while the dimer showed a slightly different activity
profile with the maximum around pH 4.7. The rate
constants for CLP (dimer) and CL (monomer) were similar
between pH 5.0 and 6.5, whereas the dimer had about
A
B
Fig. 2. SDS/PAGE analysis of Artemia cathepsin L fractions from
Mono S column. (A) Approximately 4.5 lg of Mono S fractions a–f
shown in Fig. 1 were applied to individual lanes of a 12% polyacryl-
amide gel, and following electrophoresis, the gel was stained with
Coomassieblue.Theproteaseactivityoffractionsa–fwas determined
prior to electrophoresis using the TNBS assay, and the results (pro-
tease activity per mg protein) are shown in brackets below each lane.
The migration position of CL and CLAP, the catalytic and noncata-
lytic subunits, respectively, of the protease are shown on the right,
while protein standards are shown on the left. (B) Lanes labelled e(1.5
and 3.0 lg) and f(1.5 and 3.0 lg) show the electrophoretic position of
CLAP fractions eand f, respectively, after re-chromatography on
Mono S (Fig. 1B,C). The (0) at the bottom shows the absence of
protease activity in eand fafter re-chromatography. Mw, molecular
mass marker.
Fig. 3. Isoelectric focusing of CLAP. Twenty-five micrograms of
CLAP fractions eand f(Fig. 1B,C) in a volume of 100 lLwere
applied to the top of separate glass tubes containing 6% acrylamide as
described in Materials and methods. Tubes containing pI standards
and column buffer only were prepared. After the proteins reached their
equilibrium positions, the gels containing the CLAP e,f, pI standards,
and buffer only were removed from their glass tubes, soaked in distilled
water for 5–10 min then stained with silver reagent. The pI values
assigned to bands in columns eand fwere determined from both IEF
standards (Std) and buffer control gelruninparallel.Thenumbersat
the right represent the pI values of the major bands in eand f, while the
numbers at the left are the pI values of standard proteins. The arrow at
the right represents the pI value of 6.84 calculated for the unmodified
CLAP protein based on its deduced amino acid composition.
FEBS 2004 Cathepsin L and cell adhesion protein in Artemia (Eur. J. Biochem. 271) 4017
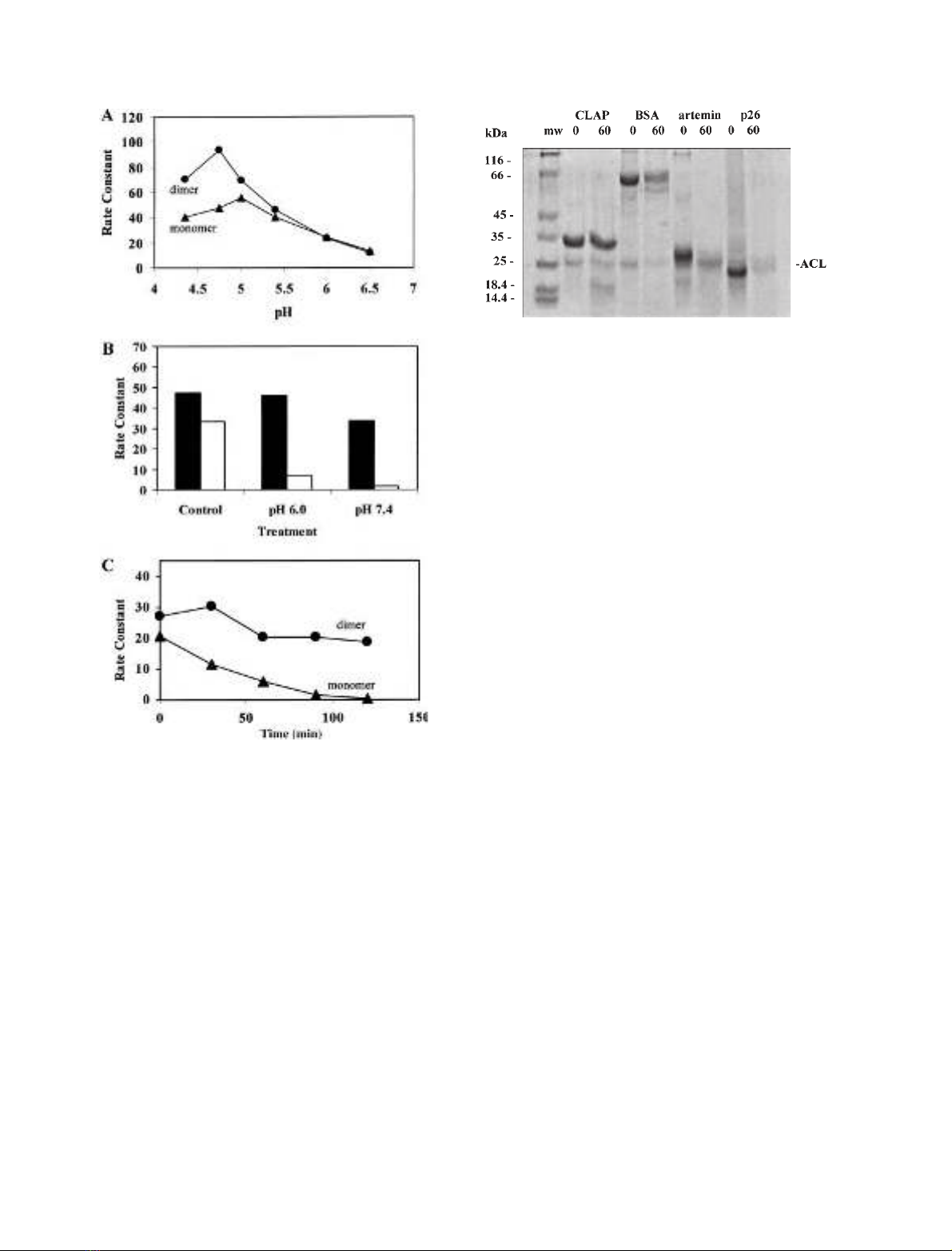
2-fold higher activity at pH 4.3–4.7. Preincubation (1 h at
30 C) of Artemia CL at pH 6.0 and 7.4 resulted in 85%
and 95% loss of cathepsin L activity, respectively, com-
pared to CLP which was less affected by these treatments
(Fig. 4B). Also, the monomer was completely inactivated
after 2 h preincubation at 40 C and pH 6.8, whereas the
dimer retained about 70% of its initial activity under these
conditions (Fig. 4C). Similar differences in cathepsin L
activity were observed at all incubation temperatures
between 40 and 53 C (data not shown). Overall, the CLP
complex is more stable than CL below pH 5, and between
pH 6.0 and 7.4 at temperatures exceeding that found in
Artemia’s natural environment [6].
Resistance of CLAP to degradation by
Artemia
cathepsin L monomer
EarlyresearchontheArtemia cysteine protease demonstra-
ted that native CLP undergoes autodegradation when
stored below pH 5 irrespective of temperature [23]. In the
present study we tested the sensitivity of CLAP and BSA,
artemin, and p26 to the Artemia CL. Results showed that
CLAP is resistant to hydrolysis by CL at 30 C and pH 5.0,
while BSA and two abundant proteins in Artemia embryos,
artemin and p26, are degraded by Artemia CL after 60 min
incubation (Fig. 5).
Abundance of the catalytic and noncatalytic subunits
of CLP at various stages in the
Artemia
life cycle
Artemia grown in the laboratory were collected at different
stages in the life cycle, and total protein isolated from
different tissues or whole animals was analyzed for the
catalytic and noncatalytic subunits using Western blotting
after SDS/PAGE separation of the proteins. Ovisacs with
encysted embryos contained the largest amount of both
protease subunits (about 0.15% of total protein) in nearly
equimolar amounts (Fig. 6). Ovisacs containing nonency-
sted embryos contained considerably less of the Artemia
CLP subunits (0.038% of the total protein in the extract),
while somatic tissues in gravid females and immature
females had still smaller amounts of each subunit
Fig. 4. Activity of the monomeric and dimeric forms of Artemia embryo
cathepsin L at different pH and temperatures. (A) CLP (dimer) and CL
(monomer) were assayed at different pH for cathepsin L activity (rate
constants). Each reaction vessel contained 40–60 pmoles of the active
protease. (B) Different forms of the protease (solid bars, CLP; unfilled
bars, CL) were incubated for 1 h at 30 Cin25m
M
KCl, 10 m
M
sodium phosphate, 10% glycerol and 0.2 mgÆmL
)1
BSA at the pH
indicated, then assayed for cathepsin L activity at pH 5.0 and 30 C
and the rate constants determined. The control represents CL
(monomer) and CLP (dimer) maintained at 0 C and pH 6.8 prior to
the assay. (C) Incubation vessels were set up to contain 80–100 pmoles
of CL (monomer) and CLP (dimer) in pH 6.8 buffer as described in
(B). The vessels were incubated at 40 C and aliquots were removed at
30 min intervals, assayed for cathepsin L activity at pH 5.0, and their
rate constants determined.
Fig. 5. Sensitivity of various proteins to Artemia cathepsin L monomer.
Reaction vessels contained 50 m
M
sodium acetate, pH 5.0, 0.5 m
M
dithiothreitol, 2.4 lg of CL (monomer), and 12–14 lg of CLAP, BSA,
artemin or p26 in a final volume of 40 lL. After 0 and 60 min incu-
bation at 30 C, 10 lL were taken from each reaction vessel for ana-
lysis by SDS/PAGE on a 12% gel. The numbers above each lane
represent the incubation time of the monomer with proteins shown
above each lane. Left lane (mw) contains molecular mass standard
proteins with their molecular mass (kDa) shown at the left. The
migration position of the Artemia cathepsin L monomer is shown at
the right (ACL). Faint bands at 16–18 kDa in the 60 min lanes rep-
resent CL autodegradation products observed in similar experiments
using Western blotting.
4018 A. H. Warner et al.(Eur. J. Biochem. 271)FEBS 2004






















![Bộ Thí Nghiệm Vi Điều Khiển: Nghiên Cứu và Ứng Dụng [A-Z]](https://cdn.tailieu.vn/images/document/thumbnail/2025/20250429/kexauxi8/135x160/10301767836127.jpg)
![Nghiên Cứu TikTok: Tác Động và Hành Vi Giới Trẻ [Mới Nhất]](https://cdn.tailieu.vn/images/document/thumbnail/2025/20250429/kexauxi8/135x160/24371767836128.jpg)


