
DtpB (YhiP) and DtpA (TppB, YdgR) are prototypical
proton-dependent peptide transporters of Escherichia coli
Daniel Harder
1
,Ju
¨rgen Stolz
1
, Fabio Casagrande
2
, Petr Obrdlik
3
, Dietmar Weitz
1
,
Dimitrios Fotiadis
2,*
and Hannelore Daniel
1
1 Molecular Nutrition Unit, Technical University of Munich, Freising, Germany
2 M.E. Mu
¨ller Institute for Structural Biology, Biozentrum, University of Basel, Switzerland
3 IonGate Biosciences GmbH, Frankfurt, Germany
Uptake of peptides into Escherichia coli cells is
thought to be mediated by three different transport
systems represented by the dipeptide permease Dpp,
the tripeptide permease TppB, and the oligopeptide
permease Opp [1–3]. Despite the fact that these pro-
teins seem to discriminate according to the backbone
length of peptide substrates, they do show some over-
lapping specificity. Although their prime physiological
role is in the uptake of peptide-bound amino acids as
an economic process to provide energy substrates and
building blocks for cellular metabolism, peptide uptake
seems also to be involved in signaling processes and
metabolic adaptation [4]. Whereas Opp and Dpp rep-
resent ATP-binding cassette transporters with periplas-
matic binding proteins [5–7], TppB belongs to the
family of proton-dependent peptide symporters that
utilize the proton gradient as driving force and lack
cognate binding proteins [8]. After identification of the
Keywords
E. coli; peptide; PTR; TppB; transport
Correspondence
H. Daniel, Molecular Nutrition Unit,
Technical University of Munich, Am Forum
5, D-85350 Freising, Germany
Fax: +49 8161713999
Tel: +49 8161713400
E-mail: daniel@wzw.tum.de
*Present address
Institute of Biochemistry and Molecular
Medicine, University of Berne, Switzerland
(Received 18 February 2008, revised 20
March 2008, accepted 23 April 2008)
doi:10.1111/j.1742-4658.2008.06477.x
The genome of Escherichia coli contains four genes assigned to the peptide
transporter (PTR) family. Of these, only tppB (ydgR) has been character-
ized, and named tripeptide permease, whereas protein functions encoded by
the yhiP,ybgH and yjdL genes have remained unknown. Here we describe
the overexpression of yhiP as a His-tagged fusion protein in E. coli and
show saturable transport of glycyl-sarcosine (Gly-Sar) with an apparent
affinity constant of 6.5 mm. Overexpression of the gene also increased the
susceptibility of cells to the toxic dipeptide alafosfalin. Transport was
strongly decreased in the presence of a protonophore but unaffected by
sodium depletion, suggesting H
+
-dependence. This was confirmed by puri-
fication of YhiP and TppB by nickel affinity chromatography and reconsti-
tution into liposomes. Both transporters showed Gly-Sar influx in the
presence of an artificial proton gradient and generated transport currents
on a chip-based sensor. Competition experiments established that YhiP
transported dipeptides and tripeptides. Western blot analysis revealed an
apparent mass of YhiP of 40 kDa. Taken together, these findings show
that yhiP encodes a protein that mediates proton-dependent electrogenic
transport of dipeptides and tripeptides with similarities to mammalian
PEPT1. On the basis of our results, we propose to rename YhiP as DtpB
(dipeptide and tripeptide permease B), by analogy with the nomenclature
in other bacteria. We also propose to rename TppB as DtpA, to better
describe its function as the first protein of the PTR family characterized in
E. coli.
Abbreviations
AMCA, b-Ala-Lys-N
e
-7-amino-4-methylcoumarin-3-acetic acid; CCCP, carbonyl cyanide m-chlorophenylhydrazone; DDM, n-dodecyl-b-D-
maltoside; Gly-Sar, glycyl-sarcosine; IPTG, isopropyl-thio-b-D-galactoside; TMPD, N,N,N¢,N¢-tetramethyl-p-phenylenediamine.
3290 FEBS Journal 275 (2008) 3290–3298 ª2008 The Authors Journal compilation ª2008 FEBS

corresponding gene ydgR [8,9], it was genetically classi-
fied as a member of the peptide transport (PTR) fam-
ily (also called proton-dependent oligopeptide
transporter or POT family). These transporters are
found essentially in all living organisms from bacteria
to humans, with examples such as DtpT from Lacto-
coccus lactis, Ptr2 from Saccharomyces cerevisiae, and
the mammalian transporters PEPT1 and PEPT2
[10,11]. The first attempts to functionally characterize
TppB in Salmonella typhimurium [12,13] and in E. coli,
employing deletion mutants [14,15], provided some
information on substrate preferences, and demon-
strated upregulation of transport upon anaerobiosis
and by Leu [13,16]. Although TppB, like Dpp, was
shown to transport dipeptides and tripeptides, it
seemed to prefer tripeptides, and in particular those of
a hydrophobic nature. The transport mode of TppB,
however, was poorly characterized. We recently cloned
ydgR encoding TppB, and overexpressed the gene for
a detailed functional analysis of the encoded trans-
porter and its initial structural characterization. We
observed a striking functional similarity between YdgR
and the mammalian PEPT1 protein [17], and therefore
the bacterial PTR transporters may serve as models
for elucidating the structure. They are easily purified in
large quantities for crystallization approaches to derive
appropriate structural models. These might then be
applied also to the human proteins, which are also of
great interest for pharmacalogical applications, e.g.
delivery of peptidomimetic drugs in the intestine.
Three of the four E. coli members of the PTR family
(yhiP,ybgH and yjdL) have not been studied yet. We
here report the cloning of yhiP and the purification
and characterization of the corresponding transport
protein. Furthermore, we compared the functional fea-
tures of YhiP with that of TppB to improve our
understanding of peptide transport processes in bacte-
ria. We established that tppB and yhiP code for proto-
typical H
+
-coupled symporter proteins that are
specific for dipeptides and tripeptides. By analogy to
similar transporters in other prokaryotes, we propose
to name them Dtp (dipeptide and tripeptide permeas-
es), with the first one identified being DtpA (former
TppB, YdgR), and DtpB (YhiP) being described here
as the second member of the PTR family in E. coli.
Results
Functional overexpression of dtpB in E. coli
Using a similar approach as previously reported for
dtpA (tppB,ydgR) [17], we amplified the dtpB (yhiP)
gene of E. coli from genomic DNA by PCR using
gene-specific primers. The gene was cloned into the
pET-21 vector fused with a C-terminal hexahistidine-
tag. The expression level of dtpB in E. coli
BL21(DE3)pLysS was assayed by western blot analysis
with an antibody against the hexahistidine-tag. This
antibody detected a protein with an apparent molecu-
lar mass of about 40 kDa in membrane preparations
of dtpB-overexpressing cells (Fig. 1A, upper panel,
lane 2). As no signal was obtained with isopropyl-thio-
b-d-galactoside (IPTG)-induced cells carrying an
empty vector (Fig. 1A, lane 1), this band clearly repre-
sented DtpB, which was also confirmed by the signal
of the nickel affinity chromatography-purified DtpB
(Fig. 1A, lane 3). The expected molecular mass based
on the amino acid sequence of DtpB is 54.6 kDa. A
similar increased mobility in SDS ⁄PAGE was observed
for DtpA, where we provided evidence that the protein
was complete by probing it with a second antibody
that recognized an added N-terminal tag [17].
To assess the function of DtpB, we determined the
uptake of radiolabeled [
14
C]glycyl-sarcosine (Gly-Sar),
a commonly used reporter substrate for mammalian
peptide transporters (Fig. 1B). Cells overexpressing
dtpB showed a higher uptake of Gly-Sar than control
cells. This was also the case for DtpA, indicating that
both proteins mediate efficient Gly-Sar uptake. As a
second approach to assess the functionality of the
transport protein, we performed growth experiments
(Fig. 1C) with the toxic phosphonopeptide alafosfalin,
a known substrate of DtpA. E. coli cells carrying the
expression vector for dtpB or the empty vector were
pregrown in the presence of a nonlethal concentration
of alafosfalin (500 lgÆmL
)1
, 2.55 mm), and gene
expression was induced by addition of IPTG. Cells
overexpressing dtpB showed strong growth inhibition
1 h after induction. The fact that cells carrying the
empty vector, noninduced cells or induced cells with-
out alafosfalin showed no growth inhibition demon-
strated the specificity of DtpB-mediated uptake of
alafosfalin. In similar experiments with DtpA, lower
alafosfalin concentrations (200 lgÆmL
)1
, 1.02 mm)
were needed for growth inhibition [17]. To directly
compare the substrate affinities of DtpB and DtpA, we
determined the uptake rates in cells overexpressing the
corresponding genes as a function of Gly-Sar concen-
tration (Fig. 1D). Transport was found to be saturable
with an apparent K
t
of 6.5 mmfor DtpB and 1 mm
for DtpA. We next investigated whether transport
requires Na
+
or H
+
(Fig. 1E). When we replaced
Na
+
in the buffer by choline, this had only a minor
effect on uptake of Gly-Sar, whereas the presence of
the proton ionophore carbonyl cyanide m-chloro-
phenylhydrazone (CCCP) caused complete inhibition
D. Harder et al. Proton-dependent peptide transporters of E. coli
FEBS Journal 275 (2008) 3290–3298 ª2008 The Authors Journal compilation ª2008 FEBS 3291
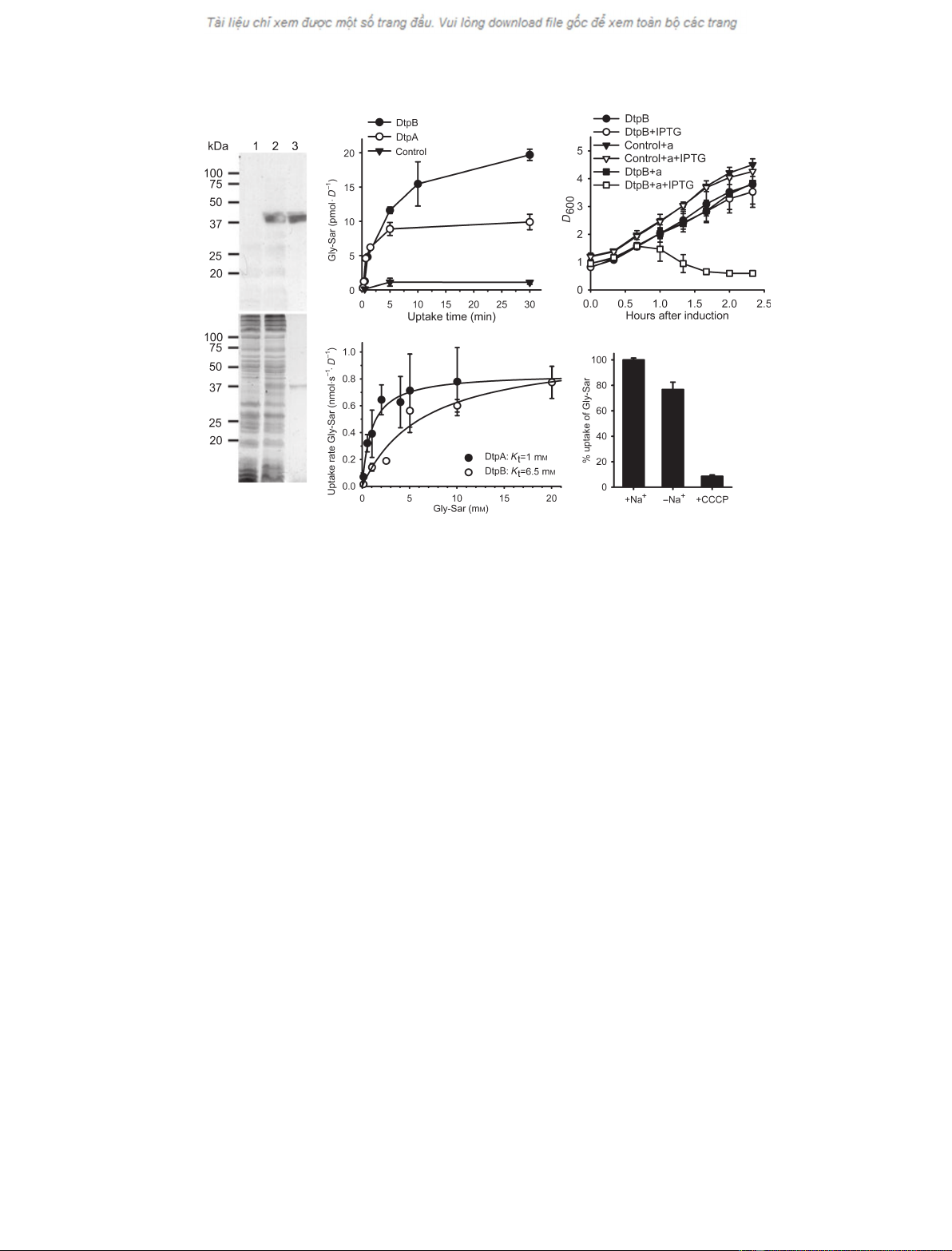
of transport. This suggests that uptake via DtpB
depends on the proton-motive force, and that DtpB
may function as a proton–peptide cotransporter.
Substrate specificity of DtpB
To assess the substrate specificity of DtpB (and DtpA,
for comparison), we performed competition experi-
ments with 1 mmGly-Sar as a substrate and 10 mm
competitors in E. coli cells overexpressing either dtpB
or dtpA. The uptake rates in the presence of the com-
petitor are presented in Table 1. The substrate specific-
ity data presented here for DtpA essentially match
those determined with b-Ala-Lys[b-Ala-Lys-N
e
-7-
amino-4-methylcoumarin-3-acetic acid(AMCA)], a fluo-
rescent dipeptide analog used as a substrate in our
previous study [17]. Similar to DtpA, neither the
amino acid Ala nor the tetrapeptide Ala-Ala-Ala-Ala
caused significant inhibition of DtpB-mediated trans-
port. In the case of DtpB, essentially all tested compet-
itors reduced transport to a lower extent than with
DtpA, which reflects its generally lower affinity.
Despite this, there were differences between DtpB and
DtpA when inhibitory effects of individual peptides
were compared. In the case of DtpB, peptides contain-
ing d-stereoisomers of l-Ala essentially failed to inhibit
uptake, whereas for DtpA, strong inhibition was
observed with d-Ala when it was placed in an N-termi-
nal position. Virtually no competition was seen in both
transporters with peptides containing only d-Ala. We
also assessed the effects of charged amino acid residues
and their spatial position on the uptake of Gly-Sar. In
the case of DtpA, a positively charged amino acid at
the N-terminus caused stronger inhibition than one
at the C-terminus. A negatively charged residue was
clearly preferred at the C-terminus. This was also
observed with DtpB, but here inhibition was higher with
Gly-Asp than in the case of DtpA. Selected b-lactam
A B C
D E
Fig. 1. Overexpression of dtpB in E. coli. (A) Anti-His-tag western blot (upper panel) and Coomassie-stained (lower panel) SDS gel (12.5%
acrylamide) of E. coli membranes of control cells (lane 1, 50 lg of total protein) or membranes containing overproduced DtpB (lane 2, 50 lg
of total protein) or purified DtpB (0.5 lg upper panel, 1.5 lg lower panel). (B) Uptake of [
14
C]Gly-Sar (1 mM)byE. coli cells carrying the
expression plasmid for dtpA or dtpB or the empty vector 3 h after induction with IPTG. Overexpression of either gene causes increased
Gly-Sar transport relative to the control. n= 2, ± SD. (C) Growth inhibition by alafosfalin. Induced (+ IPTG) or not induced cells, carrying
either the expression plasmid for dtpB or an empty plasmid, were grown in the presence of 500 lgÆmL
)1
(2.55 mM) alafosfalin (+ a) or with-
out alafosfalin. n= 2, ± SD. (D) Saturation kinetics for DtpA and DtpB. Uptake assays were performed with E. coli cells overexpressing dtpA
or dtpB, in the presence of various concentrations of Gly-Sar. The uptake velocities were determined using four time points (10, 20, 40 and
60 s). K
t
values were determined by nonlinear regression analysis of the presented data. n‡2, ± SD. (E) Sodium and proton dependence of
Gly-Sar uptake. For E. coli cells overexpressing dtpB, Gly-Sar (50 lM) uptake rates were determined from a 10 min time course. The rates
were determined in buffers containing 150 mMNaCl (+ Na
+
), or cholinechloride (150 mM) or in the presence of NaCl and the protonophore
CCCP (10 lM); 100% corresponds to 3.3 pmolÆs
)1
ÆD
)1
.n= 2, ± SD.
Proton-dependent peptide transporters of E. coli D. Harder et al.
3292 FEBS Journal 275 (2008) 3290–3298 ª2008 The Authors Journal compilation ª2008 FEBS
antibiotics (peptidomimetics) are known substrates of
mammalian peptide transporters. DtpA showed efficient
inhibition when an excess of cefadroxil, cefalexin or
cephradine was added, whereas only cephradine and
cefalexin significantly reduced Gly-Sar uptake via DtpB.
Reconstitution of DtpB and DtpA
Although transport inhibition by the protonophore
CCCP (Fig. 1E) already suggested H
+
-dependence, we
characterized the transport mode in more detail by
reconstitution in a cell-free system. Membranes of
E. coli containing the overproduced transport proteins
were fused with proteoliposomes containing bovine
cytochrome coxidase. A transmembrane proton gradi-
ent (inside negative) is generated when cytochrome c
oxidase is supplied with electrons, which are trans-
ferred from reduced ascorbate via N,N,N¢,N¢-tetram-
ethyl-p-phenylenediamine (TMPD) and cytochrome c.
Before addition of ascorbate, we observed a slow
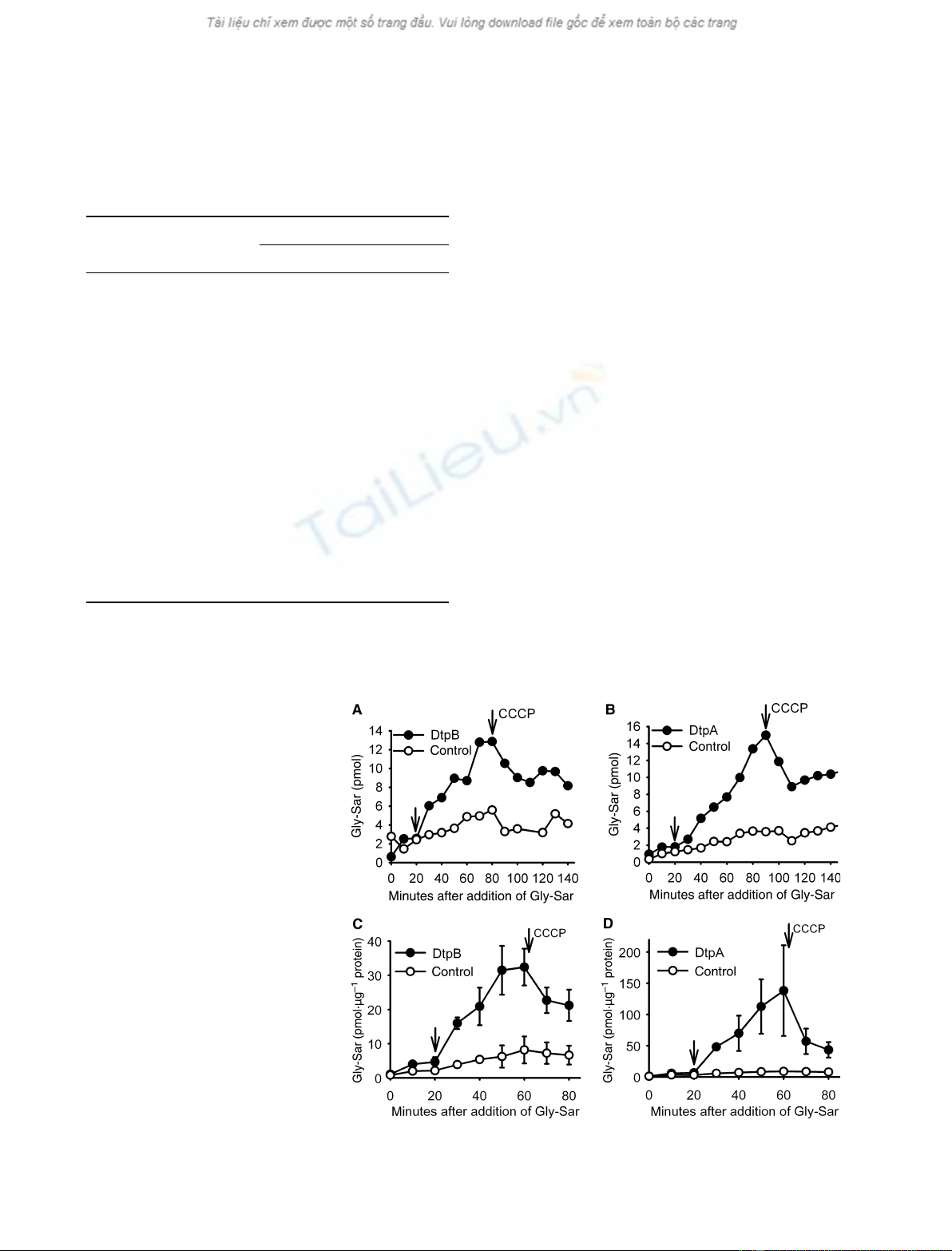
influx of Gly-Sar into all types of vesicles. Addition of
ascorbate, however, caused a marked increase in Gly-
Sar influx into vesicles containing either DtpA or
DtpB, but not into control vesicles (Fig. 2A,B). Efflux
of Gly-Sar was observed when the proton gradient was
dissipated by addition of CCCP, indicating accumula-
tion of substrate above the extravesicular concentra-
tion in the presence of the proton gradient.
Table 1. Substrate specificity of DtpA and DtpB. Uptake of Gly-
Sar in % ± standard deviation (n‡2). Competitor was present at
a concentration of 10 mM(10-fold excess) except otherwise
indicated. The uptake time was 30 s. For DtpA, 100% = 200
pmolÆs
)1
ÆD
)1
. For DtpB, 100% = 50 pmolÆs
)1
ÆD
)1
. ND, not
determined.
Competitor
[
14
C]Gly-Sar uptake (%)
DtpA DtpB
None 100 ± 19 100 ± 11
Gly-Sar 27 ± 11
a
71 ± 12
a
Gly-Sar (100 mM)ND 32±6
a
Ala 76 ± 2 124 ± 20
Ala-Ala 1 ± 0
a
17 ± 3
a
Ala-Ala-Ala 1 ± 0
a
17 ± 3
a
Ala-Ala-Ala-Ala 78 ± 22 110 ± 20
L-Ala-D-Ala 63 ± 11
a
102 ± 3
D-Ala-L-Ala 5 ± 5
a
88 ± 3
D-Ala-D-Ala 84 ± 32 102 ± 3
Gly-Lys 90 ± 40 99 ± 2
Lys-Gly 2 ± 1
a
57 ± 12
a
Gly-Asp 26 ± 6
a
17 ± 1
a
Asp-Gly 54 ± 16
a
97 ± 5
Cefamandole 145 ± 50 101 ± 18
Cefuroxime 100 ± 2 99 ± 19
Cephradine 29 ± 5
a
57 ± 4
a
Cefalexin 35 ± 7
a
78 ± 2
a
Cefadroxil 19 ± 1
a
90 ± 23
a
Significant (P< 0.05) reduction relative to assay containing no
competitor.
Fig. 2. In vitro transport studies of DtpB
and DtpA. (A) Uptake of Gly-Sar (175 lM)in
membrane vesicles generated by fusion of
membranes from dtpB-overexpressing or
control cells with cytochrome coxidase-con-
taining proteoliposomes. After 20 min
(arrow), a proton gradient was established
by addition of a mix of ascor-
bate ⁄TMPD ⁄cytochrome cand destroyed
by addition of 10 lMCCCP as indicated. (B)
As (A), but with membranes of dtpA-over-
expressing cells (one of two similar experi-
ments). (C) Uptake of Gly-Sar (175 lM)in
liposomes containing purified DtpB and
cytochrome coxidase. After 20 min (arrow),
a proton gradient was established by addi-
tion of a mix of ascorbate ⁄TMPD ⁄cyto-
chrome cand destroyed by addition of
10 lMCCCP as indicated. n= 2, ± SD. (D)
As (C), with purified DtpA instead of DtpB.
n= 2, ± SD.
D. Harder et al. Proton-dependent peptide transporters of E. coli
FEBS Journal 275 (2008) 3290–3298 ª2008 The Authors Journal compilation ª2008 FEBS 3293
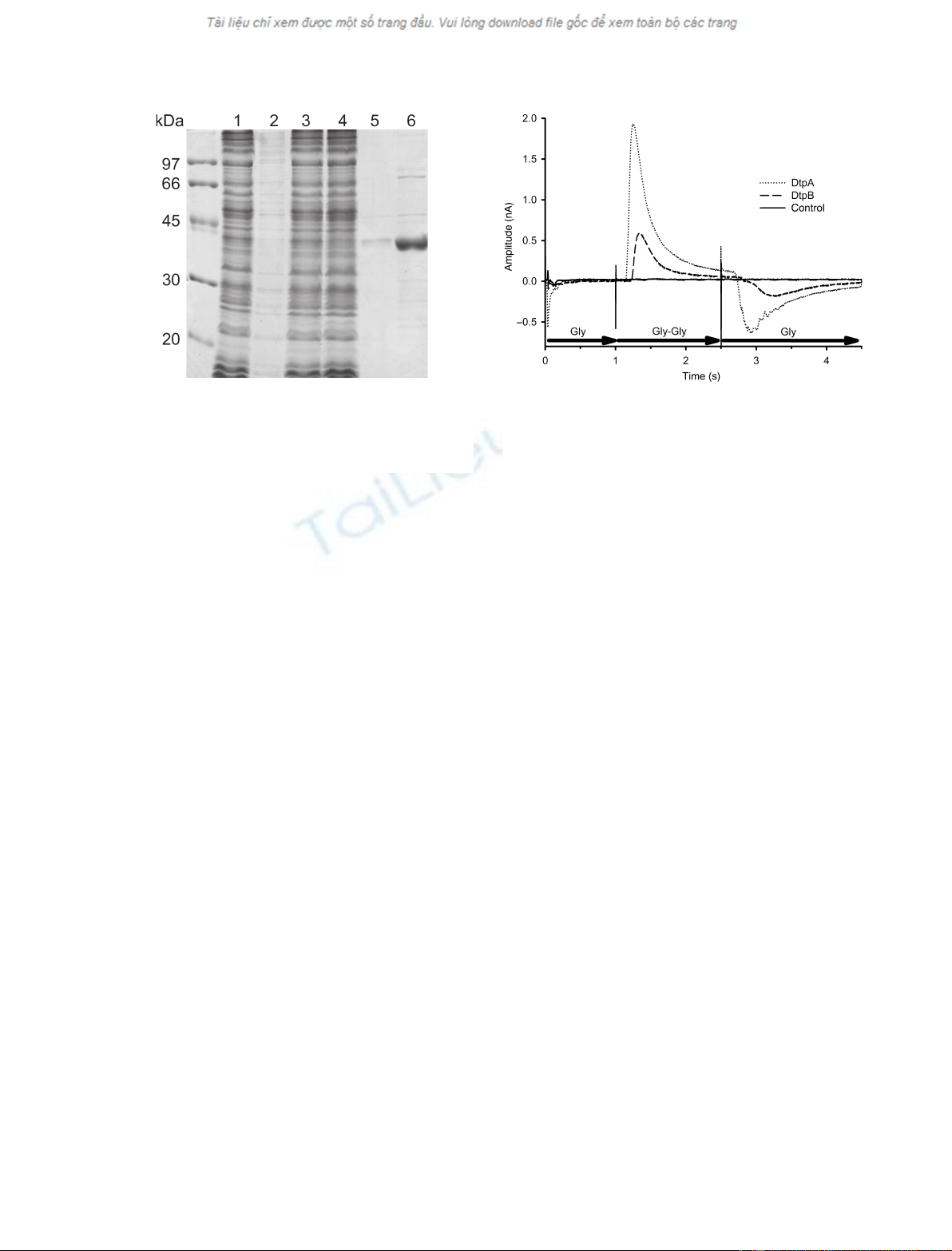
Next, we purified DtpB and DtpA from E. coli
membranes. The membranes were solubilized with the
detergent n-dodecyl-b-d-maltoside (DDM), and the
proteins were purified by metal-affinity chromato-
graphy, making use of the C-terminal His-tags. This
procedure yielded about 2–3 mg of relatively pure pro-
tein per liter of cell culture (Fig. 3). For reconstitution
into liposomes, the detergent was removed by adsorp-
tion to Bio-Beads. Subsequently, the liposomes contain-
ing the peptide transporters were fused with vesicles
containing cytochrome coxidase, using a freeze–thaw–
sonication cycle. Gly-Sar uptake after initiation of the
proton gradient increased almost 10-fold in the case of
DtpB and 60-fold in the case of DtpA (Fig. 2C,D), and
substrate efflux was observed after addition of CCCP.
To unequivocally demonstrate the electrogenic nature
of the transport process, we next used a chip-based
assay system, in which proteoliposomes containing
DtpB or DtpA were adsorbed onto a gold surface of
the SURFE
2
R
one
setup (IonGate Biosciences GmbH,
Frankfurt, Germany). This system is based on a solid-
support membrane technology, and allows detection of
capacity-coupled currents induced by movement of
charged molecules across a lipid bilayer [18,19]. After
adsorption of the proteoliposomes on the gold surface,
the SURFE
2
R
one
setup was used to let different buffers
flow for a certain time over this surface. Transport was
initiated by the exchange of a buffer containing Gly
(Gly included for charge and osmotic equilibration
only) against a buffer containing the dipeptide Gly-Gly.
Figure 4 shows that Gly-Gly induced significant cur-
rents in proteoliposomes containing DtpA or DtpB,
but not in control liposomes lacking the transport
proteins. The current spikes at the changing of the
buffer are artefacts of the working valves. The negative
current peaks observed after changing back to the Gly-
containing washing buffer most likely represent the
backflow of charges (Gly-Gly with protons) out of the
liposomes, as the substrate gradient is now reversed. As
the transport studies were performed with Na
+
-free
buffers at pH 7.0, the observed currents must originate
from proton movement coupled to dipeptide transloca-
tion in a symport mechanism. As in the cytochrome c
oxidase-energized vesicles, DtpA caused higher signals
than DtpB, suggesting that DtpA has a higher turnover
rate or higher stability in vitro.
Discussion
Despite the fact that peptide uptake in E. coli has been
studied for more than 20 years [1–3], essentially no
biochemical characterization has been performed on
any of the proteins. Another prokaryotic member of
the PTR family, DtpT from L. lactis, has been studied
in more detail, and some features of its membrane
topology are known [20]. With the focus on the four
E. coli genes that carry the typical PTR family motif
[11], we first cloned and overexpressed dtpA, and then
purified and characterized the corresponding protein
(DtpA ⁄TppB ⁄YdgR) [17]. Here we provide a detailed
functional analysis of DtpB as the second as yet
Fig. 3. Purification of DtpB. Coomassie-stained SDS gel (12.5%
acrylamide) of Ni
2+
–nitrilotriacetic acid purified DtpB. Lanes: (1)
DDM-solubilized membranes; (2) pellet after solubilization; (3)
supernatant after solubilization; (4) Ni
2+
–nitrilotriacetic acid flow-
through; (5) wash; (6) elution at about 150 mMimidazole.
Fig. 4. Electrophysiological experiment on proteoliposomes. The
electrical response of liposome-reconstituted DtpB (dashed line) or
DtpA (dotted line) to a change from a solution without substrate
(Gly) to a solution with substrate (Gly-Gly). The solid line shows the
recording from a sensor loaded with protein-free liposomes. The
current peaks indicate proton cotransport, and the negative peak is
back-flow of charges out of the liposomes (one of > 10 similar
experiments using ‡2 sensors).
Proton-dependent peptide transporters of E. coli D. Harder et al.
3294 FEBS Journal 275 (2008) 3290–3298 ª2008 The Authors Journal compilation ª2008 FEBS


























