
A multi-protein complex containing cold shock domain (Y-box)
and polypyrimidine tract binding proteins forms on the
vascular
endothelial growth factor
mRNA
Potential role in mRNA stabilization
Leeanne S. Coles
1,
*, M. Antonetta Bartley
1,
*, Andrew Bert
1
, Julie Hunter
1
, Steven Polyak
2
, Peter Diamond
1
,
Mathew A. Vadas
1,3
and Gregory J. Goodall
1,3
1
Division of Human Immunology, The Hanson Institute, Institute of Medical and Veterinary Science;
2
Division of Biochemistry,
Department of Molecular Biosciences, The University of Adelaide;
3
Department of Medicine, The University of Adelaide,
North Terrace, Adelaide, South Australia, Australia
Vascular endothelial growth factor (VEGF) is a key regu-
lator of angiogenesis and post-transcriptional regulation
plays a major role in VEGF expression. Both the 5¢-and
3¢-UTR are required for VEGF post-transcriptional regu-
lation but factors binding to functional sequences within
the 5¢-UTR have not been fully characterized. We report
here the identification of complexes, binding to the VEGF
mRNA 5¢-and3¢-UTR, that contain cold shock domain
(CSD) and polypyrimidine tract binding (PTB) RNA
binding proteins. Analysis of the CSD/PTB binding sites
revealed a potential role in VEGF mRNA stability, in both
noninduced and induced conditions, demonstrating a gen-
eral stabilizing function. Such a stabilizing mechanism had
not been reported previously for the VEGF gene. We further
found that the CSD/PTB-containing complexes are large
multiprotein complexes that are most likely preformed in
solution and we demonstrate that PTB is associated with the
VEGF mRNA in vivo. Complex formation between CSD
proteins and PTB has not been reported previously. Analysis
of the CSD/PTB RNA binding sites revealed a novel CSD
protein RNA recognition site and also demonstrated that
CSD proteins may direct the binding of CSD/PTB com-
plexes. We found the same complexes binding to an RNA-
stabilizing element of another growth factor gene, suggesting
a broader functional role for the CSD/PTB complexes.
Finally, as the VEGF gene is also regulated at the tran-
scriptional level by CSD proteins, we propose a combined
transcriptional/post-transcriptional role for these proteins in
VEGF and other growth factor gene regulation.
Keywords: cold shock domain proteins; Y-box protein;
polypyrimidine tract binding protein; mRNA stabilization;
vascular endothelial growth factor.
VEGF is an essential regulator of angiogenesis that acts on
vascular endothelial cells to induce proliferation and
promote cell migration [1–3]. Disregulated VEGF expres-
sion is implicated in a number of diseases that are
characterized by abnormal angiogenesis [1–6]. In the case
of solid tumors, the overexpression of VEGF, produced in
response to activated oncogenes, growth factors or low
oxygen conditions (hypoxia), plays a major role in promo-
ting tumor angiogenesis and progression [1–3,7]. Both the
cancer cells themselves and nontumor support cells, such as
fibroblasts, are sources of VEGF [8]. In contrast, in the case
of coronary artery disease, inadequate VEGF expression
rather than VEGF overexpression, plays a role in disease
progression. A number of cell types, including cardiac
myocytes, fibroblasts and endothelial cells produce VEGF
in response to hypoxia, but this natural response is not
sufficient to prevent the further progression of heart disease
[9–11]. It is therefore important to understand the mecha-
nisms of VEGF regulation to develop means to control
VEGF expression.
Post-transcriptional regulation plays a major role in
VEGF expression, with regulation occurring at the level of
splicing, mRNA stability and translation [2,7]. The VEGF
mRNA is normally unstable and its stability is increased in
response to cytokines and stress conditions such as hypoxia
[7,11–14]. Regions in both the 5¢-and3¢-UTR have been
shown to be involved in VEGF mRNA stabilization
[7,12,13,15–18]. The presence of an internal ribosome entry
site (IRES) in the VEGF 5¢-UTR ensures continual
translation of the VEGF mRNA in stress conditions that
normally decrease cap-dependent translation [19–21]. Little
is known about the factors involved in VEGF post-
transcriptional regulation. Factors such as HuR and
hnRNPL have been implicated in hypoxic stability via their
Correspondence to L. S. Coles, Division of Human Immunology,
The Hanson Institute, Institute of Medical and Veterinary Science,
Frome Road., Adelaide, South Australia, 5000, Australia.
Fax: + 61 88 2324092, Tel.: + 61 88 2223432,
E-mail: leeanne.coles@imvs.sa.gov.au
Abbreviations: CSD, cold shock domain; IRES, internal ribosome
entry site; VEGF, vascular endothelial growth factor.
*These authors contributed equally to this work.
(Received 16 October 2003, revised 14 December 2003,
accepted 16 December 2003)
Eur. J. Biochem. 271, 648–660 (2004) FEBS 2004 doi:10.1111/j.1432-1033.2003.03968.x
actions on the VEGF 3¢-UTR [17,18] but factors involved in
stability or translational regulation have not been identified
on the 5¢-UTR.
The single-strand RNA and DNA binding, cold shock
domain (CSD) (also known as Y-box) proteins, play
diverse roles in both transcriptional and post-transcrip-
tional regulation of growth factor and stress response
genes [22–29]. CSD proteins have several family members
which are defined by the presence of a central highly
conserved 70 amino acid region called the cold shock
domain [24,25,29]. The central domain is required for
sequence-specific RNA binding, while the adjacent
C-terminal domain has a more nonspecific role in
stabilizing binding [24–27]. There are two types of
nongerm cell CSD proteins and these are called dbpB
(also known as YB-1, MSY-1, chkYB-1b, EF1A, p50 and
FRGY1) and dbpA (MSY4, chkYB-2 and YB2/RYBa).
DbpB and dbpA CSD proteins are ubiquitously expressed
and are highly conserved across species. Highly conserved
germ cell-specific CSD proteins also exist such as MSY-2
and FRGY2 [22–25,29]. In addition there are CSD-related
proteins such as UNR (upstream of N-ras) which contains
multiple conserved CSD domains [30]. CSD proteins
stabilize growth factor/stress response mRNAs in response
to stress signals [31–33] and also act as general mRNA
stabilizers [34–37]. In addition, CSD and CSD-related
proteins have been shown to play a role in cap-dependent
and [26,27,38–42] IRES-dependent [43–46] translation and
in RNA splicing [47,48]. In the case of the GM-CSF
(granulocyte-macrophage colony stimulating factor)
growth factor gene, CSD proteins have been shown to
play a combined role at both the transcriptional and post-
transcriptional levels [22,23,49,50]. As we have recently
shown a role for CSD proteins in regulation of the VEGF
gene at the transcriptional level [51], and given the diverse
functions of CSD proteins, relevant to VEGF expression,
we investigated a role for CSD proteins in VEGF post-
transcriptional regulation.
We now show here that CSD proteins can bind to both
the 5¢-and3¢-UTR of the VEGF mRNA. We find that
CSD proteins form a cytoplasmic complex on VEGF
mRNA that also contains the multifunctional single-
strand RNA/DNA binding protein, PTB [43–46,52–57],
and that the binding of this complex may be involved in
general VEGF mRNA stabilization. The CSD/PTB-con-
taining cytoplasmic complex also forms on a stability
element in the interleukin-2 (IL-2) 5¢-UTR suggesting a
similar mechanism of regulation of stability of growth
factor mRNAs.
Materials and methods
Plasmid constructs
The pGEM44, pGEM46 and pGEM47 constructs were
generated by cloning segments of the mouse VEGF
5¢-UTR, that were amplified by PCR from the pfVEGF
construct [15], into pGEM4Z (Promega). The pGEM44,
46 and 47 constructs contain, respectively, mouse VEGF
5¢-UTR sequences +1 to +325, +461 to +727 and +735 to
+1014 (relative to the transcription start site at +1) [58]
(Fig. 1). The pGEMV1 construct, containing the VEGF
5¢-UTR CSD site 1 sequences (+150 to +185) was
constructed by cloning double strand oligonucleotides (with
EcoRI 5¢-andHindIII 3¢-ends) into pGEM4Z. pGEMV37,
39, 15, 17 and 19 were similarly constructed, except that they
contained mutant versions of the CSD site 1 sequences
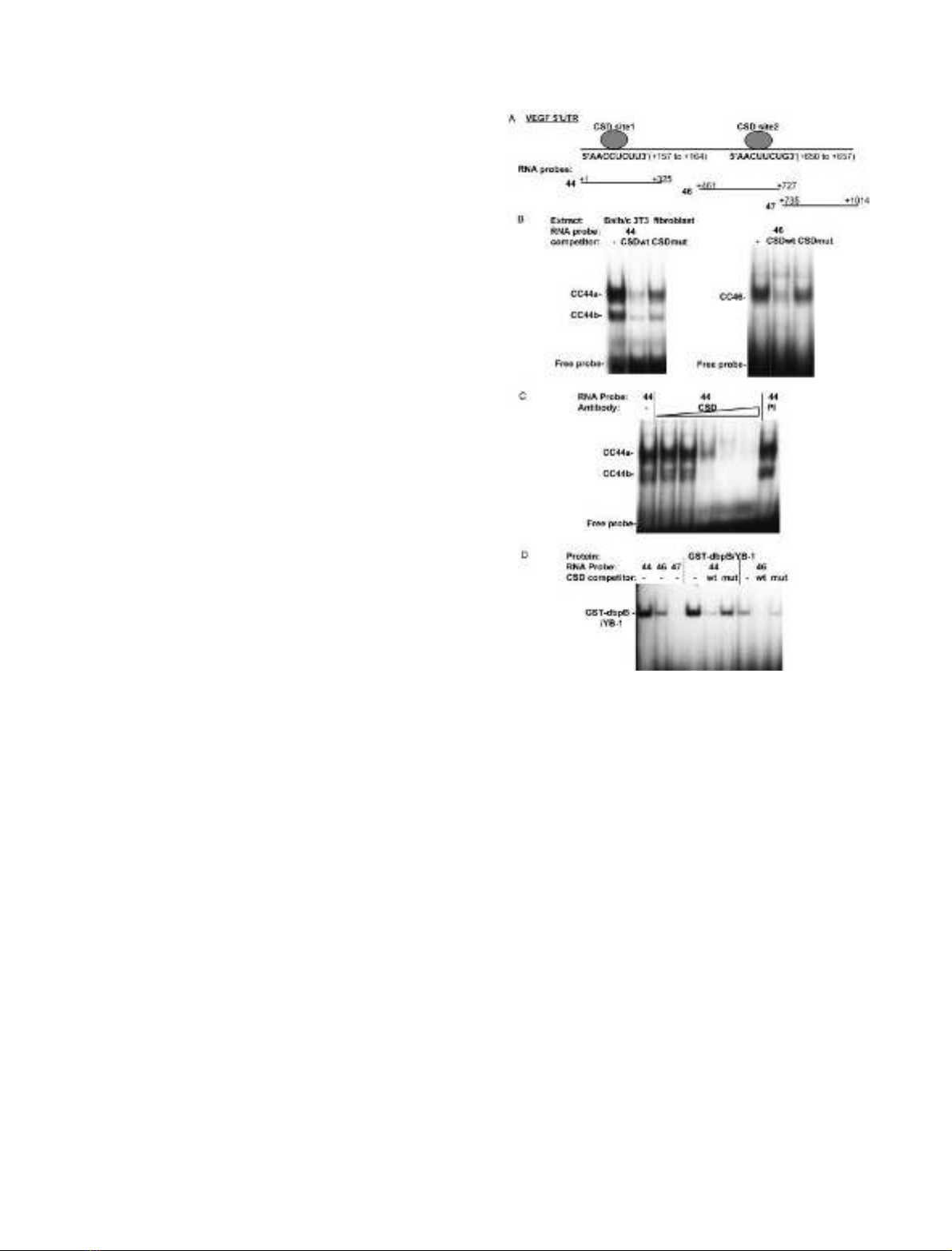
Fig. 1. The VEGF 5¢-UTR binds cytoplasmic and recombinant CSD
proteins. (A) Schematic of the mouse VEGF 5¢-UTR. The sequences
and coordinates (relative to the mRNA start site +1) [58] of consensus
CSD binding sites (CSD site 1,2) are indicated. The coordinates of
RNA probe sequences are also indicated. RNA probes were derived
from pGEM44, pGEM46 and pGEM47, respectively. (B) Balb/c 3T3
fibroblast cytoplasmic extracts were incubated without competitor (–)
or with unlabeled single-strand DNA competitor oligonucleotides
containing wild-type (CSDwt) and mutant (CSDmut) CSD binding
sites [49–51].
32
P-Labeled RNA probes (44 and 46) were then imme-
diately added and RNase T1 digested complexes analyzed by gel shift
assay. Cytoplasmic complexes CC44a, CC44b and CC46 and unbound
RNA probe are indicated. (C) Cytoplasmic extracts were preincubated
with anti-CSD polyclonal Ig (CSD), with preimmune serum (PI) or left
untreated (–), followed by addition of the labeled VEGF 44 RNA
probe in a gel shift assay. Increasing amounts of anti-CSD Ig were
added. Pre-immune sera was used at the maximal concentration used
for the anti-CSD Ig. Cytoplasmic complexes CC44a and CC44b are
indicated. (D) Recombinant GST-dbpB/YB-1 was incubated with
labeled 44, 46 and 47 RNA probes. Complexes were competed with
wild-type (CSDwt) or mutant (CSDmut) CSD binding site single-
strand DNA competitors or left untreated (–).
FEBS 2004 CSD and PTB protein complexes on the VEGF mRNA (Eur. J. Biochem. 271) 649
(Figs 2 and 3). pGEMV25 and pGEMV27 were construc-
ted by cloning wild-type and mutant double strand oligo-
nucleotides containing the IL-2 5¢-UTR +1 to +35
sequences [31] (Fig. 4). pGEMVC1 was constructed by
cloning double strand oligonucleotides containing the
VEGF 3¢-UTR CSD site 3 (+1712 to +1747, relative to
the stop codon at +1, of the mouse VEGF 3¢-UTR) [59]
into pGEM4Z. pGEMVC2 and VC3 contained mutations
in the CSD site 3 sequence (Fig. 6).
The pfVEGF construct contained a reconstructed, tagged
VEGF cDNA sequence [15], composed of the entire VEGF
mouse 5¢-UTR (+1 to +1014), the coding region for the 164
aminoacidformofVEGFandtheVEGF3¢-UTR (+4 to
+2195). The major polyadenylation site is at +1861 [59]. A
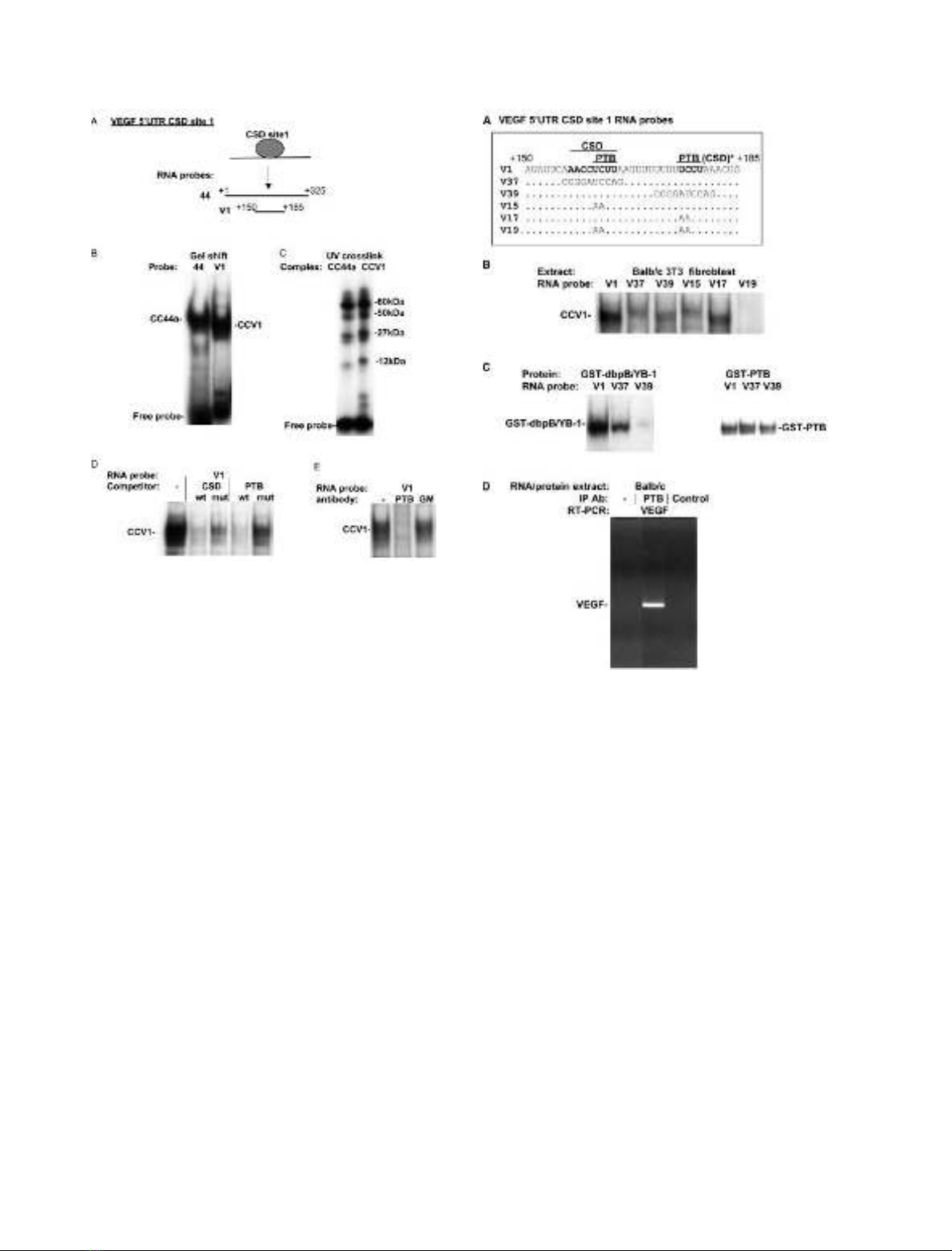
Fig. 2. The VEGF 5¢-UTR CSD-containing cytoplasmic complexes also
contain PTB. (A) Schematic of the VEGF 5¢-UTR CSD site 1. The
coordinates for the VEGF RNA probes 44 and V1 are indicated rel-
ativetothestartoftheVEGF mRNA (+1) [58]. (B) Balb/c 3T3
fibroblast cytoplasmic extract was incubated with labeled VEGF 44 or
V1 RNA probes, followed by RNase T1 digestion in a gel shift assay.
The 44 and V1 RNA probes are derived from pGEM44 and
pGEMV1, respectively. Cytoplasmic complexes CC44a and CCV1
and unbound RNA probe are indicated. (C) Cytoplasmic complexes
CC44 and CCV1, in gel shift assay gels, were exposed to UV light to
cross-link proteins in each complex to RNA. Cross-linked proteins
were then analyzed by SDS/PAGE and the sizes of cross-linked pro-
teins were calculated by subtraction of the molecular weight of bound
RNA probe. The sizes of cross-linked proteins are indicated. Cross-
link analysis is not quantitative as different proteins will crosslink to
different extents. (D) Cytoplasmic extracts were incubated with unlabe-
led wild-type or mutant CSD (CSDwt, mut) or PTB (PTB wt, mut) [52]
binding site single-strand DNA oligonucleotides, or left untreated (–).
Labeled V1 RNA probe was then immediately added and complexes
analyzed in a gel shift assay. The CCV1 complex is indicated. (E)
Cytoplasmic extracts were preincubated with an anti-PTB monoclonal
antibody (PTB), with a control anti-GM-CSF monoclonal antibody
(GM), or without antibody (–). Labeled V1 RNA probe was then
added and complexes analyzed in a gel shift assay.
Fig. 3. Sequence requirement for VEGF 5¢-UTR CCV1 CSD/PTB
complex formation-PTB complexes form on the VEGF mRNA in vivo.
(A) The sequence of the VEGF 5¢-UTR V1 RNA probe is shown and
consensus CSD and PTB protein binding site sequences are indicated.
The 3¢consensus PTB site is also found, in this report, to bind
recombinant CSD protein (labeled CSD*). The sequences of mutant
RNA probes are given under the V1 sequence. Only those bases that
are changed in the mutant probes are indicated. The RNA probes were
generated from pGEMV1, V37, V39, V15, V17 and V19 constructs,
respectively. (B) Balb/c 3T3 fibroblast cytoplasmic extracts were
incubated with labeled wild-type (V1) and mutant VEGF 5¢-UTR CSD
site 1 RNA probes in a gel shift assay. The CCV1 cytoplasmic complex
is indicated. (C) Recombinant GST-dbpB/YB-1 and GST-PTB were
incubated with labeled wild-type (V1) and mutant (V37, V39) VEGF
5¢-UTR RNA probes in a gel shift assay. The recombinant protein
complexes are indicated. (D) PTB binding to VEGF mRNA in vivo was
investigated using an RNA immunoprecipitation assay. Cytoplasmic
RNA/protein complexes (prepared in the presence of RNase inhibi-
tors) were immunoprecipitated with anti-PTB monoclonal Ig (PTB),
with an IgG2 isotype control (control), or without antibody (–). VEGF
mRNA in RNA extracted from immunoprecipitated complexes was
detected by RT-PCR. The VEGF PCR product is indicated.
650 L. S. Coles et al. (Eur. J. Biochem. 271)FEBS 2004
polylinker is positioned between the coding region and the
3¢-UTR sequences to distinguish pfVEGF mRNA from
endogenous VEGF mRNA. pfVEGFdel contains deletions
of the site 1, 2 and 3 CSD sites (Fig. 7). The sequences +156
to +179 (CSD site 1) and +650 to +666 (CSD site 2) of the
5¢-UTR were deleted and the sequences +1727 to +1740
(CSD site 3) of the 3¢-UTR were deleted.
Construction of the expression vector producing recom-
binant GST-dbpB/YB-1 (pGEXBT) has been described
previously [49,51]. pGEXPTB, for production of recombin-
ant GST-PTB, was constructed by cloning of a 1.6kb EcoRI
fragment from pcDNA3PTB (gift from T. Cooper, Baylor
College of Medicine, Houston, TX, USA), coding for
human PTB, into pGEX4T-2.
Oligonucleotides
Oligonucleotides for cloning into pGEM4Z and for use as
competitors in gel shift assays were synthesized by Gene-
works (Adelaide, Australia) and purified from nondenatur-
ing polyacrylamide gels. Single-strand oligonucleotides for
competition of CSD protein-containing complexes were
from the human granulocyte-macrophage-colony stimula-
ting factor (GM-CSF)gene.Thewild-type(CSDwt)and
mutant sequences (CSD site mutant; CSDmut) have been
described previously (GM- and GMm23-, respectively)
[49–51]. The CSD wild-type sequence binds both dbpA and
dbpB CSD proteins. The wild-type (PTBwt) and mutant
PTB (PTBmut) competitor single-strand DNA oligonucleo-
tides are from the transferrin gene (DR1 sense and DR1
sense mut1, respectively) [52].
RNA probe preparation
32
P-labeled RNA probes for gel shift analysis or RNase
protection assays were generated by in vitro transcription
from linearized plasmid templates (pGEM4Z constructs)
using SP6 (for pGEM44,46,47) or T7 (for pGEMV1, V25
and VC1) RNA polymerase (Promega) and [
32
P]UTP[aP].
Probes for RNase protection assays were processed as
previously described [15]. Probes for gel shift assays were
purified from nondenaturing polyacrylamide gels and eluted
into RNase-free water at 56 C.
Preparation of recombinant and cytoplasmic proteins
The Escherichia coli strain MC1061 transformed with
pGEXBT or pGEXPTB was induced with isopropyl thio-
b-
D
-galactoside to produce recombinant GST-dbpB/YB-1
and GST-PTB [49,51]. The fusion proteins for gel shift
analysis were purified on glutathione-Sepharose beads
(Promega). Cytoplasmic extracts were produced according
to the method of Schrieber et al. [60].
FPLC gel filtration of cytoplasmic extracts
Cytoplasmic extract from Balb/c 3T3 fibroblasts was
applied at a flow rate of 0.35 mLÆmin
)1
to a Superdex 200
column (10 mm diameter, 20 mL bed volume) pre-equili-
brated with buffer containing 150 m
M
KCl, 20 m
M
Tris/
HCl pH 7.6, 20% glycerol, 1.5 m
M
MgCl
2
,2m
M
dithio-
threitol, 0.4 m
M
phenylmethanesulfonyl fluoride and 1 m
M
Na
3
VO
4
. The CCV1 complex was eluted with the same
buffer and 0.5 mL fractions collected. The molecular mass
of the complex was estimated from the column by
comparison with the elution volumes of c-globulin, bovine
serum albumin, ovalbumin, myoglobin and vitamin B12.
Antibodies
The anti-CSD antibody is a rabbit polyclonal Ig raised
against a peptide conserved in dbpA and dbpB/YB-1 CSD
proteins across species [49,51]. The anti-PTB Ig is a mouse
monoclonal antibody (BB7; gift from D. Black, UCLA,
Los Angeles, CA, USA). A mouse monoclonal anti-
(GM-CSF) Ig (gift from A. Lopez, Hanson Institute,
IMVS, Adelaide, Australia) and an IgG2 monoclonal
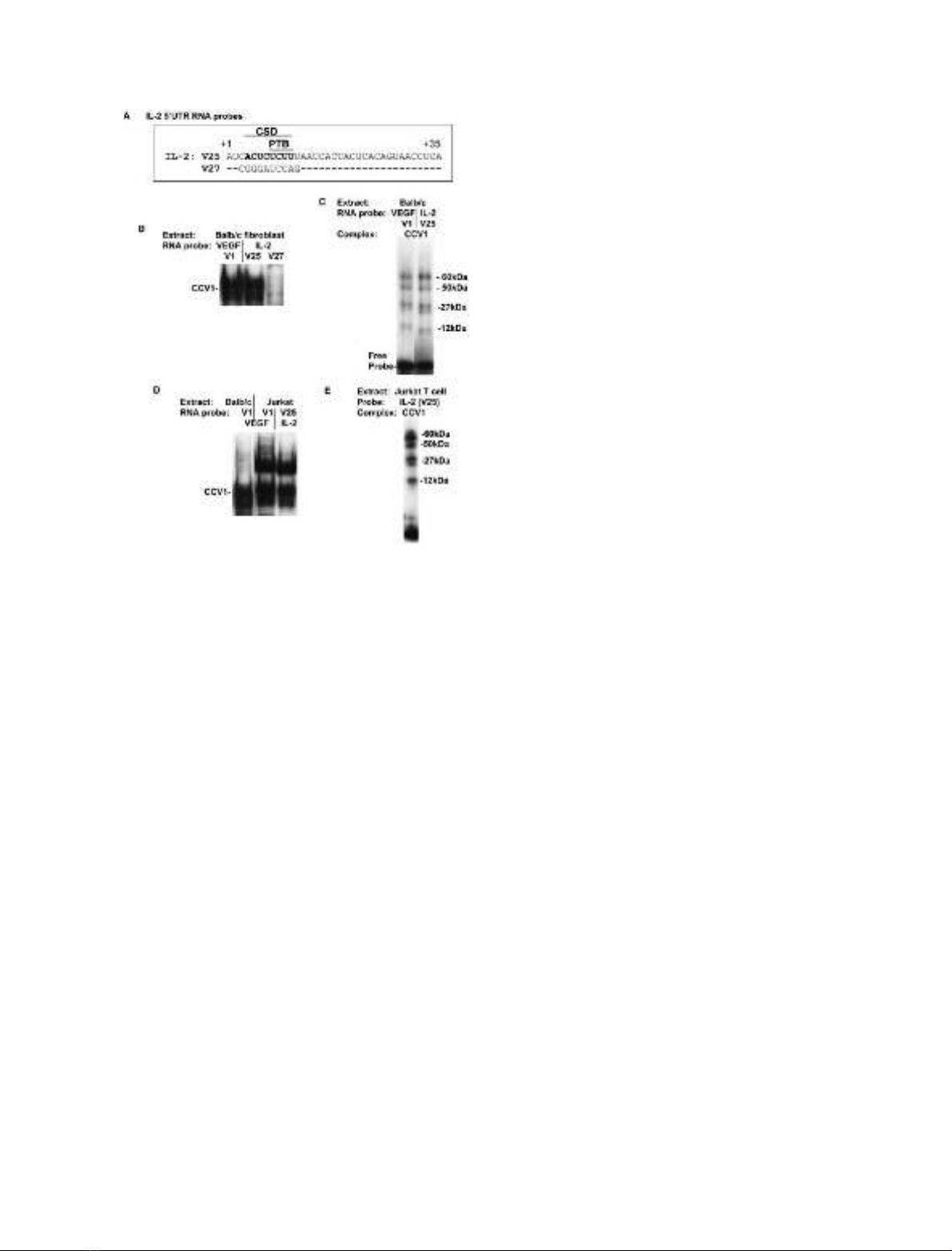
Fig. 4. CCV1 complex formation on the IL-2 5¢-UTR stability element
in fibroblasts and Jurkat T cells. (A) Sequence of the IL-2 5¢-UTR wild-
type probe V25 with consensus CSD and PTB sites indicated. The
region +1 to +22 is involved in IL-2 mRNA stabilization in T cells
[31]. The V27 mutant sequence is shown with only those bases differing
from the wild-type sequence shown. (B) Balb/c 3T3 fibroblast cyto-
plasmic extracts were incubated with labeled VEGF (V1) and IL-2
(V25,V27) 5¢-UTR RNA probes, and analyzed by gel shift. The CCV1
cytoplasmic complex is indicated. (C) The Balb/c 3T3 CCV1 com-
plexes binding to the VEGF (V1) and IL-2 (V25) RNA probes (after
RNase T1 digestion) were exposed to UV light, in gel shift gels, to
cross-link proteins to RNA. Cross-linked proteins were sized by SDS/
PAGE. The sizes of cross-linked proteins were calculated by subtrac-
tion of molecular masses of bound RNA probes. (D) Balb/c 3T3
fibroblast and Jurkat T cell cytoplasmic extracts were incubated with
labeled VEGF (V1) and IL-2 (V25) RNA probes, digested with RNase
T1 and analyzed in a gel shift assay. The CCV1 cytoplasmic complex is
indicated. (E) The Jurkat T cell CCV1 cytoplasmic complex binding to
the IL-2 V25 RNA probe was analyzed by UV cross-link analysis as
described above. The sizes of cross-linked proteins are indicated.
FEBS 2004 CSD and PTB protein complexes on the VEGF mRNA (Eur. J. Biochem. 271) 651

antibody isotype control (Silensus, Boronia, Victoria, Aus-
tralia) were used as controls for the anti-PTB antibody in gel
shift assays and RNA immunoprecipitations, respectively.
RNA gel shift analysis, competitions and antibody
analysis
RNA gel shifts were performed using
32
P-labeled RNA
probes in a 10 lL reaction mix of 0.5·TM buffer [49–51]
containing 200 m
M
KCl, 1 lg poly(dI.dC), 100 ng tRNA,
1lg bovine serum albumin and either 1 lg cytoplasmic
extract or 25 ng recombinant protein (GST-dbpB or GST-
PTB). Reactions were incubated at 4 C for 20 min,
followed by treatment with or without RNase T1 (Worth-
ington Biochemical Corp., NJ, USA) and analyzed on
6% nondenaturing polyacrylamide gels. Competition with
single-strand DNA oligonucleotides was performed by
addition of protein and 50 ng of unlabeled probe, followed
by immediate addition of the
32
P-labeled RNA probe.
Antibody blocking experiments were performed by incuba-
ting protein and antibody for 5 min before adding the
32
P-labeled probe. Antibodies did not degrade RNA probes
under the gel shift conditions used.
UV cross-linking
Cytoplasmic extracts were bound to
32
P-labeled RNA
probes in a 25 lLgelshiftreactionandfractionatedona
6% polyacrylamide gel as described above. The gel was
exposed to UV light (340 nm) for 15 min and retarded
complexes were excised after exposure overnight to X-ray
film. Protein in excised bands was analyzed by 12% SDS/
PAGE as described previously [49–51].
RNA immunoprecipitation assay
Balb/c 3T3 fibroblast extracts were prepared, as described
above, in the presence of RNase inhibitors (Promega) and
incubated with or without anti-PTB monoclonal Ig or with
an IgG2 isotype control for 60 min. RNase inhibitors were
required to prevent the loss of RNA from extracts. Protein
A sepharose CL-4B (Pharmacia, Biosciences, Uppsala,
Sweden) was added and further incubated for 60 min.
Sepharose was extracted for bound RNA (TRIzol
reagent, Invitrogen). RNA was reverse transcribed using
Superscript II (Promega) and a PCR assay for VEGF
cDNA was performed using oligonucleotides from the
mouse VEGF cDNA of 5¢-CACAGACTCGCGTTGCA-3¢
and 5¢-TGGGTGGGTGTGTCTAC-3¢. PCR products
were analyzed by agarose gel electrophoresis. The VEGF
PCR product is approximately 400 bp.
Cell culture, stable transfection and cell stimulation
Mouse Balb/c 3T3 fibroblasts and rat C6 glioma cells were
grown in Dulbecco’s modified Eagle’s medium with 10%
fetal bovine serum. Jurkat T cells were cultured in RPMI
media with 10% fetal bovine serum. For cytoplasmic
extracts, cells were grown in normoxic conditions (normal
oxygen; 20% O
2
). For the production of stably transfected
cell lines, C6 glioma cells were transfected with linearized
pfVEGF or pfVEGfdel plasmids using lipofectamineTM
2000 (Gibco BRL Life Technologies, Melbourne, Australia)
according to the manufacturer’s directions. Cells were
grown for 24–48 h and selected in 400 lgÆmL
)1
G418 [15].
Serum stimulation of stably transfected cell lines was as
described previously [15]. Hypoxic conditions (1% O
2
)were
generated in a hypoxic chamber (Edwards Instrument
Company, Sydney, Australia).
Analysis of mRNA stability
in vivo
Stable transfectants (pfVEGF or pfVEGFdel) were serum
stimulated (time 0) and concurrently incubated under
normoxic or hypoxic conditions for 1, 1.5, 2, 3 or 4 h.
Serum stimulation provides a brief pulse of transcription
from the c-fos promoter in pfVEGF/pfVEGFdel con-
structs, allowing subsequent degradation of the mRNA to
be monitored as previously described by us in analysis of the
pfVEGF construct [15]. This system allows determination
of mRNA stability directly, rather than using indirect means
such as nonspecific inhibitors of transcription. RNA was
isolated from treated cells using TRIzol
r
reagent (Invitro-
gen) according to the manufacturers instructions, and
pfVEGF/pfVEGFdel mRNA was detected by RNase pro-
tection analysis using a
32
P-labeled transcript covering the
polylinker sequence in the pfVEGF/pfVEGFdel constructs
as previously described [15]. Neomycin phosphotransferase
(neo) mRNA expressed from pfVEGF/pfVEGFdel con-
structs was detected as described [15]. Protected RNAs were
separated on denaturing polyacrylamide gels and the
amounts of specific
32
P-labeled protected pfVEGF/pfVEGF-
del or neo mRNAs were quantitated by PhosphoImager
analysis (Molecular Dynamics, Sunnyvale, CA, USA).
Levels of pfVEGF/pfVEGFdel mRNA (with time 0 levels
subtracted) were normalized with respect to the levels of neo
mRNA at each time point.
Results
The VEGF 5¢-UTR binds cytoplasmic and recombinant
CSD proteins
Sequence specific RNA binding sites for CSD proteins
have been determined in a few genes but a consensus
sequence has not been established. Analysis of the prota-
mine 1 (Prm1) 3¢-UTR has revealed a preferred binding site
of 5¢-U/C/A–C/A–C–A–U/C–C–A/C/U-3¢for mouse CSD
proteins [38–40]. This sequence is consistent with a prefer-
red sequence for Xenopus CSD proteins (FRGY1/2) of
5¢-AACAUCU-3¢[61] and with a 5¢-ACCACC-3¢sequence
from the Rous Sarcoma virus LTR that binds chicken CSD
proteins [41].
Given a potential role for CSD proteins in VEGF post-
transcriptional regulation, the VEGF 5¢-UTR was exam-
ined for CSD protein binding sites. Two potential sites at
+157 and +650 were observed. These were named CSD site
1 and CSD site 2 and have sequences of 5¢-AACCU
CU-3¢and 5¢-AACUUCU-3¢, respectively (Fig. 1A). No other
potential CSD protein binding sequences were observed.
To determine if the VEGF 5¢-UTR could bind cytoplas-
mic CSD complexes,
32
P-labeled RNA probes 44 (+1 to
+325) and 46 (+461 to +727) covering the potential CSD
sites (Fig. 1A) were bound to cytoplasmic extracts from
652 L. S. Coles et al. (Eur. J. Biochem. 271)FEBS 2004


























