Effect of ecdysone receptor gene switch ligands on
endogenous gene expression in 293 cells
Siva K. Panguluri
1
, Bing Li
2
*, Robert E. Hormann
2
and Subba R. Palli
1
1 Department of Entomology, College of Agriculture, University of Kentucky, Lexington, KY, USA
2 Intrexon Corporation, Norristown, PA, USA
Gene therapy is used to correct a defect in the expres-
sion of a gene by transferring a gene expression cas-
sette containing a promoter, a terminator, and the
coding region of a gene whose absence or defect causes
a disease. Current technology uses constitutive promot-
ers, such as the cytomegalovirus promoter, for expres-
sion of transgenes. Such an ‘always on’ arrangement is
not desirable because it can exacerbate pleiotropic
effects and also leaves no option for remediation in the
event of medical complications due to transgene
expression. To address this issue, regulated expression
of the additive or corrective gene becomes attractive
for various gene therapy applications. Despite extra
complexity, the regulated expression system can
Keywords
diacylhydrazine; ecdysone; gene therapy;
microarray; RSL-1
Correspondence
S. R. Palli, Department of Entomology,
College of Agriculture, University of
Kentucky, Lexington, KY 40546 USA
Fax: +1 859 323 1120
Tel: +1 859 257 4962
E-mail: rpalli@uky.edu
*Present address
MicroBiotiX, Inc., Worcester, MA, USA
(Received 24 June 2007, revised 13 August
2007, accepted 4 September 2007)
doi:10.1111/j.1742-4658.2007.06089.x
Regulated gene expression may substantially enhance gene therapy. Corre-
lated with structural differences between insect ecdysteroids and mamma-
lian steroids, the ecdysteroids appear to have a benign pharmacology
without adversely interfering with mammalian signaling systems. Conse-
quently, the ecdysone receptor-based gene switches are attractive for appli-
cation in medicine. In the present study, the effect of inducers of ecdysone
receptor switches on the expression of endogenous genes in HEK 293 cells
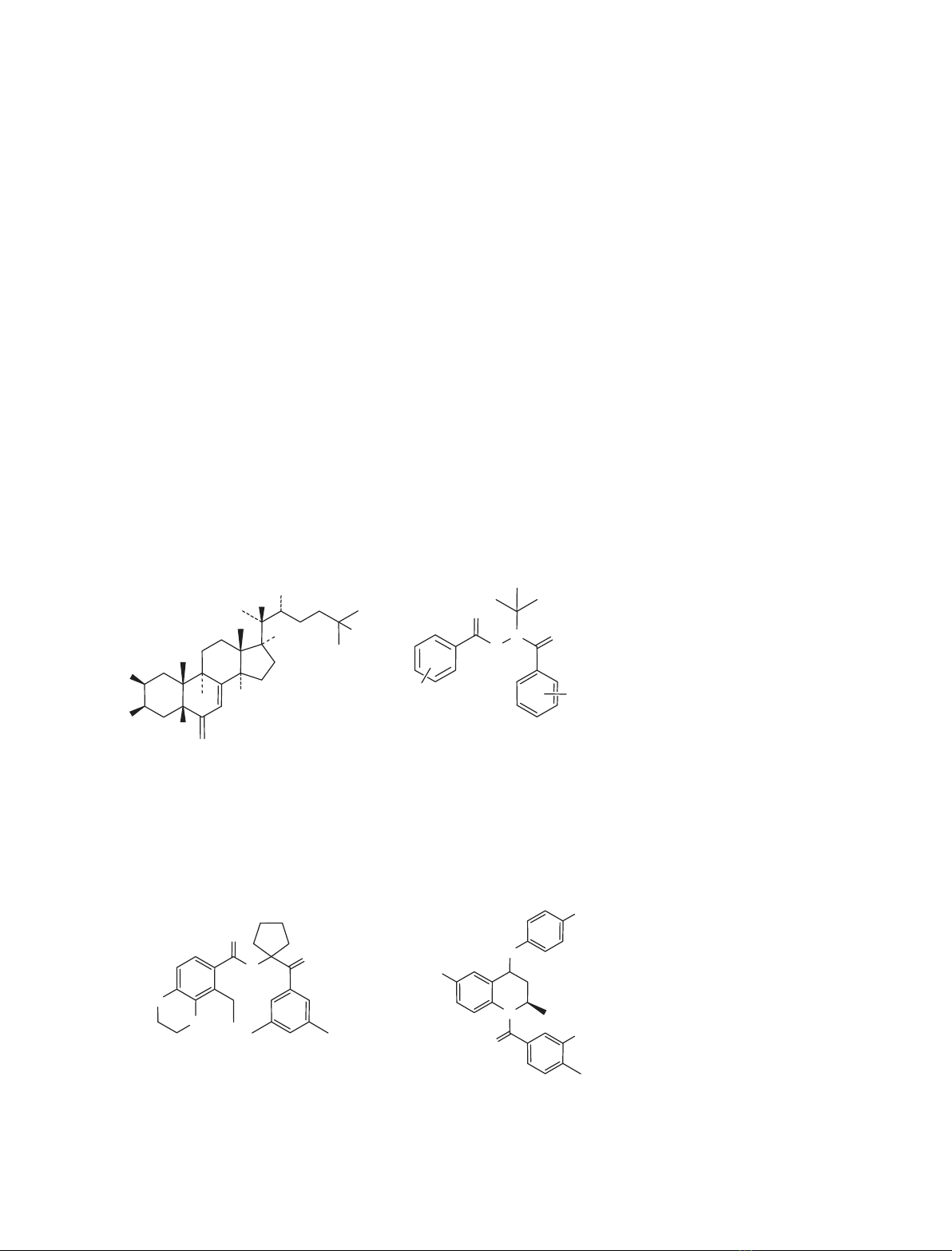
was determined. Four ligand chemotypes, represented by a tetrahydroquin-
oline (RG-120499), one amidoketone (RG-121150), two ecdysteroids
[20-hydroxyecdysone (20E) and ponasterone A (Pon A)], and four diacyl-
hydrazines (RG-102240, RG-102277, RG-102398 and RG-100864), were
tested in HEK 293 cells. The cells were exposed to ligands at concentra-
tions of 1 lm(RG-120499) or 10 lm(all others) for 72 h and the total
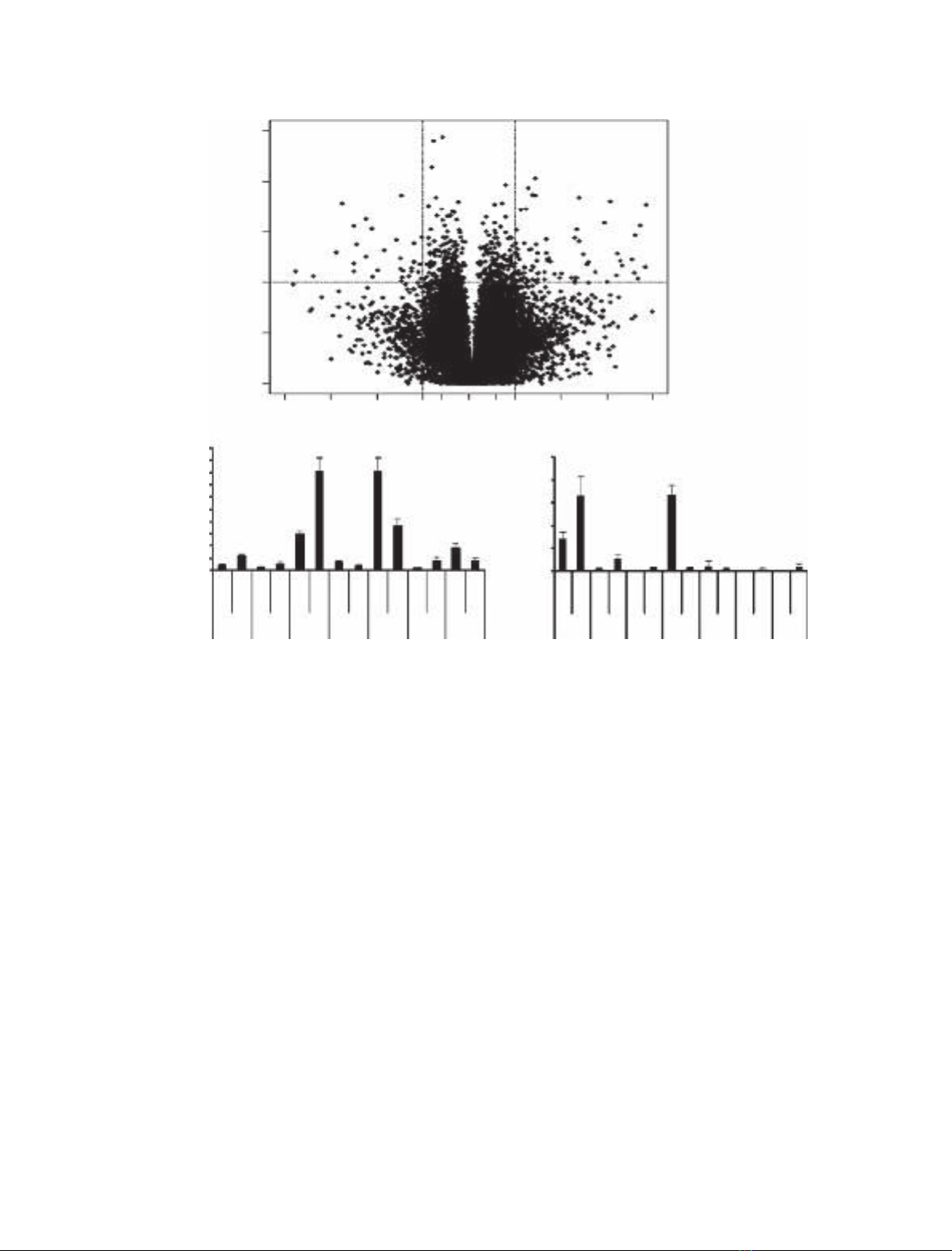
RNA was isolated and analyzed using microarrays. Microarray data
showed that the tetrahydroquinoline ligand, RG-120499 caused cell death
at concentrations ‡10 lm.At1lm, this ligand caused changes in the
expression of genes such as TNF,MAF,Rab and Reprimo.At10lm, the
amidoketone, RG-121150, induced changes in the expression of genes such
as v-jun,FBJ and EGR, but was otherwise noninterfering. Of the two ste-
roids tested, 20E did not affect gene expression, but Pon A caused some
changes in the expression of endogenous genes. At lower concentrations
pharmacologically relevant for gene therapy, intrinsic gene expression
effects of ecdysteroids and amidoketones may actually be insignificant.
A fortiori, even at 10 lm, the four diacylhydrazine ligands did not cause
significant changes in expression of endogenous genes in 293 cells and
therefore should have minimum pleiotropic effects when used as ligands
for the ecdysone receptor gene switch.
Abbreviations
AMK, amidoketone; qRT-PCR, quantitative real-time reverse transcription PCR; STAT 6, signal transducer and activator of transcription 6;
THQ, tetrahydroquinoline.
FEBS Journal 274 (2007) 5669–5689 ª2007 The Authors Journal compilation ª2007 FEBS 5669