An Optogenetic Upgrade for the Tet-OFF System
Konrad M€uller,
1
Matias D. Zurbriggen,
1,2
Wilfried Weber
1,2
1
Faculty of Biology, University of Freiburg, Sch€anzlestrasse 1, 79104 Freiburg, Germany
2
BIOSS Centre for Biological Signalling Studies, University of Freiburg, Sch€anzlestrasse
18, 79104 Freiburg, Germany; telephone: þ49 761 203 97654; fax: þ49 761 203 97660;
e-mail: wilfried.weber@biologie.uni-freiburg.de
Abstract: The rapid development of mammalian optogenetics has
produced an expanding number of gene switches that can be
controlled with the unprecedented spatiotemporal resolution of
light. However, in the “pre-optogenetic” era many networks, cell
lines and transgenic organisms have been engineered that rely on
chemically-inducible transgene expression systems but would
benefit from the advantages of the traceless inducer light. To open
the possibility for the effortless upgrade of such systems from
chemical inducers to light, we capitalized on the specific Med25VBD
inhibitor of the VP16/VP64 transactivation domain. In a first step,
we demonstrated the efficiency and selectivity of Med25VBD in the
inhibition of VP16/VP64-based transgene expression systems.
Then, we fused the inhibitor to the blue light-responsive B-LID
degron and optimized the performance of this construct with regard
to the number of Med25VBD repeats. This approach resulted in an
optogenetic upgrade of the popular Tet-OFF (TetR-VP64, tetO
7
-
P
hCMVmin
) system that allows tunable, blue light-inducible trans-
gene expression in HEK-293T cells.
Biotechnol. Bioeng. 2015;9999: 1–5.
ß2014 Wiley Periodicals, Inc.
KEYWORDS: optogenetics; inducible expression; gene switch;
TET system
In the past five years, optogenetic tools have evolved from the use of
light-triggered ion channels in neurosciences to a diverse, rapidly
expanding toolbox for broader applications in biological research
(Gautier et al., 2014; Weitzman and Hahn, 2014). Of note, several
light-responsive gene switches have been developed for mammalian
systems that respond to distinct regions of the light spectrum
(Muller et al., 2014b) and can be combined to independently control
several genes within a single cell (Muller et al., 2013, 2014a).
However, in the “pre-optogenetic” era numerous chemically-
inducible gene switches have been developed and applied to
construct gene networks, cell lines and transgenic animals (Weber
and Fussenegger, 2011). For instance, the prototype of chemically
controlled transgene expression systems, the Tet-OFF system
(Gossen and Bujard, 1992), has been adopted in thousands of
research projects, optimized Tet-OFF cell lines are offered for sale
and a wide selection of Tet-transgenic mice is available from public
repositories (Schonig et al., 2010). While the introduction of the Tet-
OFF system (and of other chemically inducible gene switches) has
had a big impact on biological research, particularly in the rise of
mammalian synthetic biology, many of the existing networks, cell
lines and transgenic animals would be improved, if gene expression
could not only be controlled in time by the addition or removal of
tetracycline or its analogue doxycycline, but also in space. This
however, cannot be achieved by chemically controlled gene switches
due to diffusion of the inducer and even temporal control is limited
by the time needed for the inducer to diffuse in or out of the target
cells. While light offers both, extremely high temporal and spatial
resolution, it is cumbersome to start from scratch and integrate
optogenetic gene switches into existing systems that have already
been tediously optimized. Therefore, we aimed to engineer an
optogenetic upgrade for the Tet-OFF system that would open up the
possibility for the effortless control of existing Tet-OFF based
networks, cell lines or animals with light.
The Tet-OFF system is based on the tetracycline-dependent
binding of a fusion protein, consisting of the TetR-DNA binding
protein and a transactivation domain, to its operator site. The
originally published Tet-OFF system uses the VP16 transactivation
domain from Herpes simplex virus (Gossen and Bujard, 1992), but
improved versions of the system use more concise activation
domains, such as a tetrameric repeat of the minimal VP16 motif
termed “VP64” (Seipel et al., 1992). Since the VP16 transactivation
domain is also employed by many other widely-used gene switches,
we focused on this component for the design of our optogenetic
upgrade. It has been shown that VP16 induces gene expression by
recruiting the subunit 25 of the Mediator complex (Med25) (Yang
et al., 2004) and the structure of the protein domain that interacts
with VP16 has been resolved and was termed the Med25 VP16-
binding domain (Med25VBD) (Milbradt et al., 2011). Notably, it
was demonstrated that overexpressed Med25VBD binds to and
competitively inhibits VP16 in a dominant negative manner.
Conflict of Interest: None.
Contract grant sponsor: European Community’s Seventh Framework Programme
Contract grant number: FP7/2007-2013
Contract grant sponsor: ERC
Contract grant number: 259043-CompBioMat
Contract grant sponsor: Excellence Initiative of the German Federal and State
Governments
Contract grant number: EXC 294
Received 7 November 2014; Revision received 28 January 2015; Accepted 5 February
2015
Accepted manuscript online xx Month 2015;
Article first published online in Wiley Online Library
(wileyonlinelibrary.com).
DOI 10.1002/bit.25562
COMMUNICATION TO THE EDITOR
ß2014 Wiley Periodicals, Inc. Biotechnology and Bioengineering, Vol. 9999, No. xxx, 2015 1
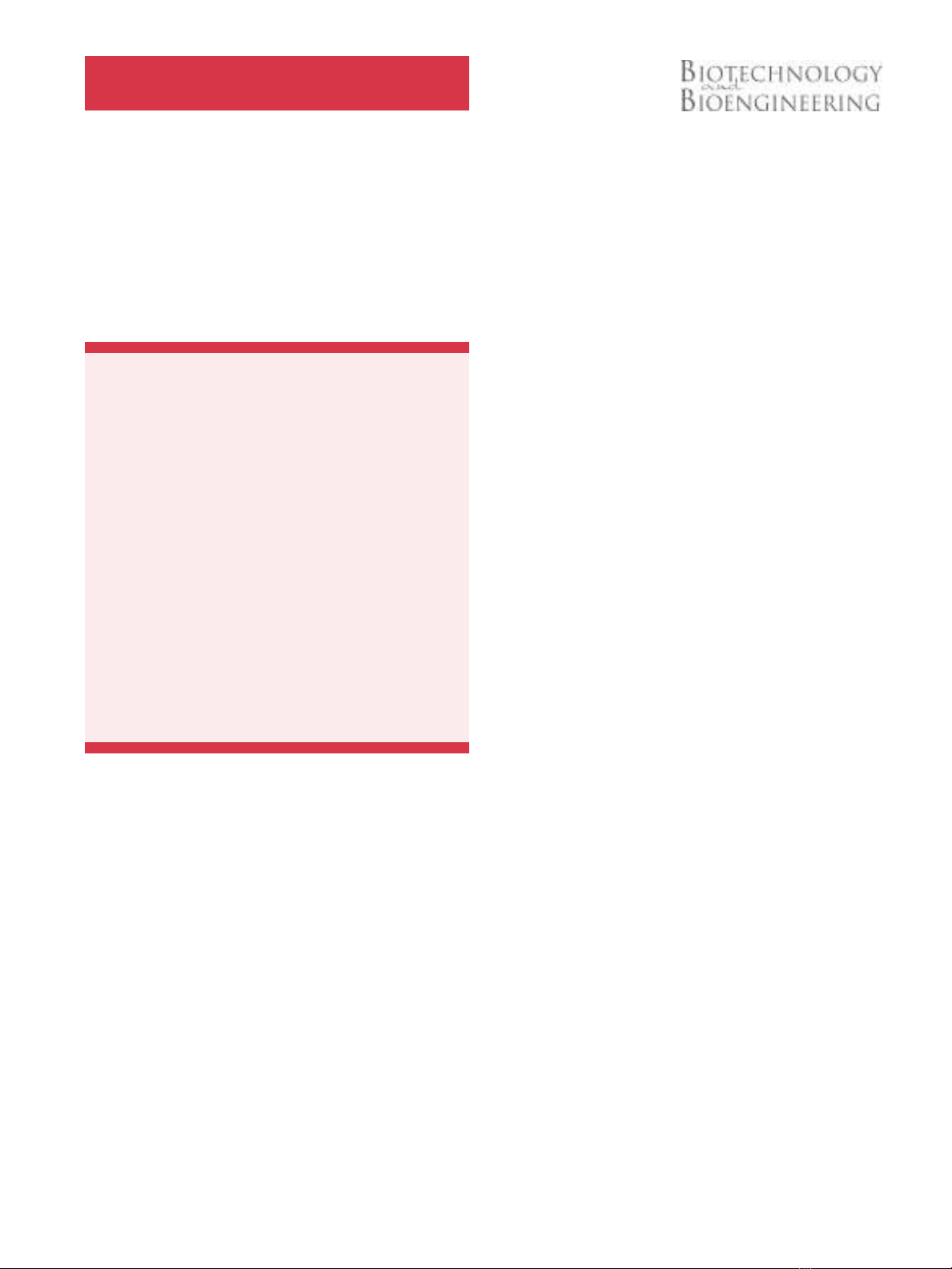
Following a systematic approach to construct a Med25VBD-
based optogenetic upgrade, we first determined the efficiency of
Med25VBD-mediated inhibition of several VP16-based transgene
expression systems. To this end, we co-transfected HEK-293T cells
with the Tet-OFF system (Gossen and Bujard, 1992), as well as with
gene-switches that are controlled by macrolide antibiotics (E-OFF;
Weber et al., 2002), streptogramin antibiotics (PIP-OFF; Fusse-
negger et al., 2000), the apple metabolite phloretin (Phloretin-OFF;
Gitzinger et al., 2009) or by tryptophan (TRP-ON; Bacchus et al.,
2012), along with increasing amounts of a nuclear-targeted fusion
protein of EGFP (as an indicator for expression) and Med25VBD.
When transfected in equal amounts (w:w) we observed a
Med25VBD-dependent repression of gene expression between 6-
fold (PIP-OFF) and 19-fold (Tet-OFF) that could be increased to a
23-fold (PIP-OFF) to 327-fold (Tet-OFF) inhibition, when
Med25VBD was transfected in 10-fold excess over the other
components (Figure 1 and Supplementary Figure S1).
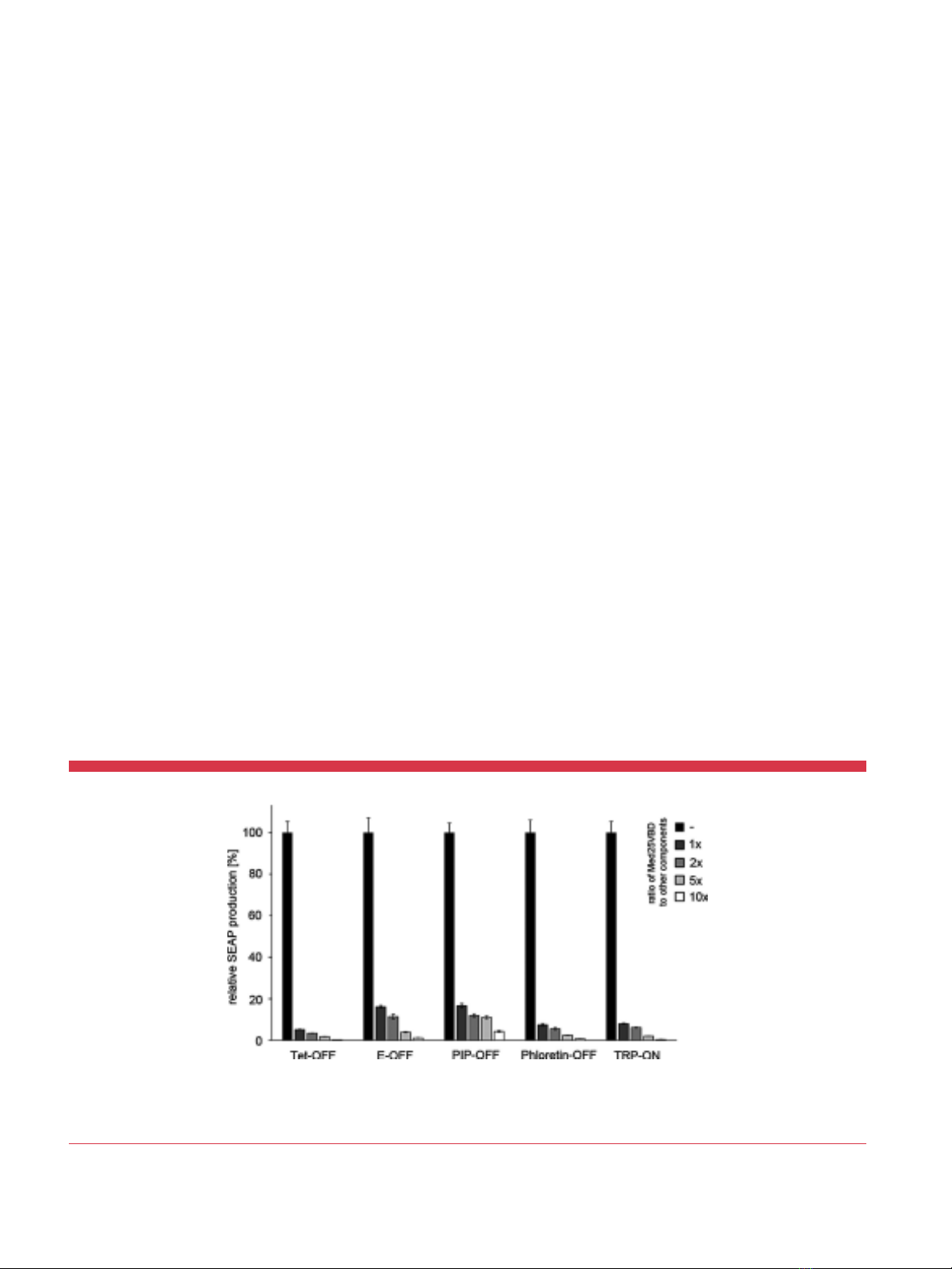
Besides the Herpes simplex virus-derived VP16 transactivation
domain and its derivatives such as the “VP64” transactivation
domain that has been constructed as a tetrameric repeat of the
minimal VP16 transactivation motif (Beerli et al., 1998; Seipel et al.,
1992), the human p65 transcriptional activation domain from NF-
kB has been used in the design of several transgene expression
systems (Urlinger et al., 2000). Unlike the VP16-transactivation
domain, p65 does not induce gene expression by recruiting a
member of the Mediator complex, but interacts with the TAF4b
subunit of the general transcription factor TFIID (Yamit-Hezi and
Dikstein, 1998; Yamit-Hezi et al., 2000), suggesting that this
transactivator is not inhibited by Med25VBD. To demonstrate that
Med25VBD not only potently represses VP16-dependent gene
expression but is also highly specific for this transactivation
domain, we transfected HEK-293T cells with versions of the Tet-
OFF system that use the VP16, VP64 or p65 transactivation
domains to induce transgene expression, along with increasing
amounts of nuclear-targeted EGFP-Med25VBD. While we already
observed strong inhibition of reporter expression from the VP16-
and VP64-based systems when Med25VBD was transfected in equal
amounts to the other components (10-fold and 20-fold respec-
tively), the p65-based system was barely repressed 2-fold at a 5-fold
excess of Med25VBD. Notably an 85-fold/229-fold repression of the
other systems was achieved under these conditions (Figure 2 and
Supplementary Figure 2).
Having confirmed that Med25VBD can be used to potently and
selectively repress VP64-dependent transgene expression, we
embarked upon the engineering of a light-controlled VP64
inhibitor. To this end, we turned to the recently published B-LID
(blue light-inducible degradation) domain (Bonger et al., 2014).
This optogenetic protein degradation tool is based on the LOV2
domain from Avena sativa phototropin 1 that has been modified by
adding an RRRG-degradation signal to its Ja-helix. In the dark, the
Ja-helix is docked to the core of the LOV2 domain, thus
sequestering the degradation signal that becomes accessible only
upon blue light illumination to induce protein degradation (Bonger
et al., 2014). To construct the optogenetic upgrade for the Tet-OFF
(TetR-VP64; tetO
7
-P
hCMVmin
-reporter) system, we fused B-LID to
the nuclear targeted EGFP-Med25VBD fusion protein. In the dark,
Med25VBD binds to VP64, inhibiting transgene expression. Only
upon blue (465 nm) light-induced exposure of the degradation
signal, breakdown of the inhibitor is induced, freeing VP64 to
induce gene expression from the minimal promoter (Figure 3A). To
optimize the inhibition of VP64 in the dark as well as the induction
of gene expression in blue light, we tested several constructs that
harbored between one and four repeats of Med25VBD between
EGFP and B-LID. Quantification of reporter expression from HEK-
293T cells transfected with the Tet-OFF system and the inhibitors,
after 24 h of illumination with 465 nm light, revealed that three
repeats of Med25VBD yielded the best (7-fold) induction of gene
expression in blue light-treated compared to dark-incubated cells
(Figure 3B). Increasing the amount of the inhibitor relative to the
components of the Tet-OFF system did not result in an improved
Figure 1.Inhibition of VP16-based expression systems by Med25VBD. HEK-293T cells were transfected with VP16-based transgene expression systems along with increasing
amounts of pKM254 coding for nuclear-localized EGFP-Med25VBD, or with an empty plasmid backbone (pRSet, black bars). Twenty-four hours post-transfection, production of the
SEAP reporter was quantified. For each group the mean SD (n¼4) is displayed relative to the respective control group without Med25VBD (black bars). Tet-OFF, pSAM200/
pMF111; E-OFF, pWW035/pWW037; PIP-OFF, pMF156/pMF172; Phloretin-OFF, pMG011/pKM502; TRP-ON, pWB024/pLMK116.
2Biotechnology and Bioengineering, Vol. 9999, No. xxx, 2015
induction of gene expression (Supplementary Table S1). Moving
beyond the Tet-OFF system, we demonstrated that other VP64-
based expression systems can also be rendered light-sensitive with
induction ratios ranging from 2-fold to 5-fold using the optimized
optogenetic upgrade (Supplementary Table S2). Since many
applications require the precise adjustment of gene expression,
we also demonstrated that the blue light-induced activity of the Tet-
OFF system with the optogenetic upgrade can be readily adjusted by
controlling the light intensity. To this end, HEK-293T cells that had
been transfected with the Tet-OFF system and the optimized light-
controlled VP64-inhibitor were illuminated with 465 nm light of
increasing intensities. The quantification of reporter production
revealed an intensity-dependent increase of reporter levels that
reached saturation at an intensity of about 8 mmol m
2
s
1
(Figure 3C). Finally, we compared the ON and OFF kinetics of the
Tet-OFF system with the optogenetic upgrade to those of the basic
Tet-OFF system. To study the induction of gene expression, HEK-
293T cells were transfected with the Tet-OFF system with or without
the optimized light-responsive inhibitor and cultivated in the OFF
state (i.e., in the dark or in the presence of tetracycline,
respectively). Next, the systems were switched to the ON state by
illumination with 465 nm light (Tet-OFF with optogenetic upgrade)
or by the withdrawal of tetracycline (basic Tet-OFF system). The
reporter production was monitored for 23 h and revealed
comparable kinetics for both systems (Figure 3D). The kinetics
of the shut-off of gene expression were analyzed by illuminating
cells transfected with the Tet-OFF system and the optogenetic
upgrade with activating 465 nm light for 6 h. Then, gene expression
was terminated either by displacing TetR-VP64 from the response
construct through the addition of tetracycline, or by moving the
cells to the dark. Control cells were illuminated with 465 nm light or
incubated in the dark for the entire experiment. The analysis of
reporter production revealed that the addition of tetracycline
resulted in a rapid termination of gene expression (Figure 3E and
Supplementary Figure S3), while the expression shut-off by moving
the cells to the dark was delayed (Figure 3E). The delayed
termination of expression is most likely caused by the requirement
for the de novo synthesis of the inhibitor to return the
optogenetically upgraded Tet-OFF system to the OFF state.
In conclusion, we have developed an optogenetic upgrade for the
VP64-based Tet-OFF system that allows the rapid and adjustable
induction of gene expression upon blue light illumination. In light
of the multitude of synthetic networks, cell lines and organisms that
have been constructed based on the Tet-OFF system, we expect that
our tool will allow researchers to upgrade their existing systems
with minimal effort to substitute the inducer tetracycline with blue
light, and thus profit from its unprecedented spatiotemporal
resolution. The overexpression of Med25VBD could possibly
interfere with certain endogenous transactivators that target this
component of the transcription initiation complex (Yang et al.,
2004). Still, we have demonstrated the selective inhibition of VP16/
VP64 by Med25VBD with respect to the p65 transactivation
domain. Thus opening up the possibility to selectively regulate the
VP16/VP64 transactivators with light in synthetic gene networks,
without affecting modules that utilize the p65 transactivation
domain.
Materials and Methods
DNA Cloning
The construction of expression vectors is given in detail in
Supplementary Table S3.
Cell Culture and Transfection
Human embryonic kidney fibroblasts (HEK-293T; Mitta et al.,
2002) were maintained in Dulbecco’s modified Eagle’s medium
(PAN, cat. no. P03-0710) supplemented with 10% FBS (PAN, cat. no.
P30-3602, batch no. P101003TC), 100 U mL
1
of penicillin and
0.1 mg mL
1
of streptomycin (PAN). Where indicated, tetracycline
was added to the culture medium to a final concentration of
3mg mL
1
from a 3 mg mL
1
stock in EtOH. Cells were transfected,
using an optimized polyethylene-imine-based method (PEI, linear,
MW: 25 kDa) (Polyscience) (Muller et al., 2014c). In brief,
1 mg mL
1
PEI solution in H
2
O was adjusted to pH 7.0 with HCl,
sterile filtered and stored at 80C in aliquots. Next, 70,000 cells
were seeded per well of a 24-well plate and cultivated overnight.
Aliquots of 0.75 mg of DNA were diluted in 50 mL of OptiMEM
(Invitrogen) and mixed with 2.5 mL of PEI solution in 50 mL of
OptiMEM by vortexing (amounts scaled to one well). After 15 min
incubation at room temperature, the precipitate was added to the
cells. The culture medium was replaced 5 h after the transfection.
Unless indicated, all plasmids were transfected in equal
amounts (w:w).
Illumination Conditions
Blue (465 nm) light illumination was performed by custom-made
LED arrays (Muller et al., 2014c). Light intensity was adjusted using
Figure 2.Specificity of Med25VBD. HEK-293T cells were transfected with three
versions of the Tet-OFF system constituted by TetR-VP16 (pSAM200), TetR-VP64
(pKM551) or TetR-p65 (pKM568) and a SEAP reporter plasmid (pMF111). Along with the
tetracycline-controlled transgene expression systems, increasing amounts of nuclear-
targeted EGFP-Med25VBD (pKM254) or an empty plasmid backbone (pRSet, black
bars) were transfected. Twenty-four hours post-transfection, SEAP production was
quantified in the cell culture medium. For each group the mean SD (n¼4) is
displayed relative to the respective control group without Med25VBD (black bars).
M€
uller et al.: An Optogenetic Upgrade for the Tet-OFF 3
Biotechnology and Bioengineering 3
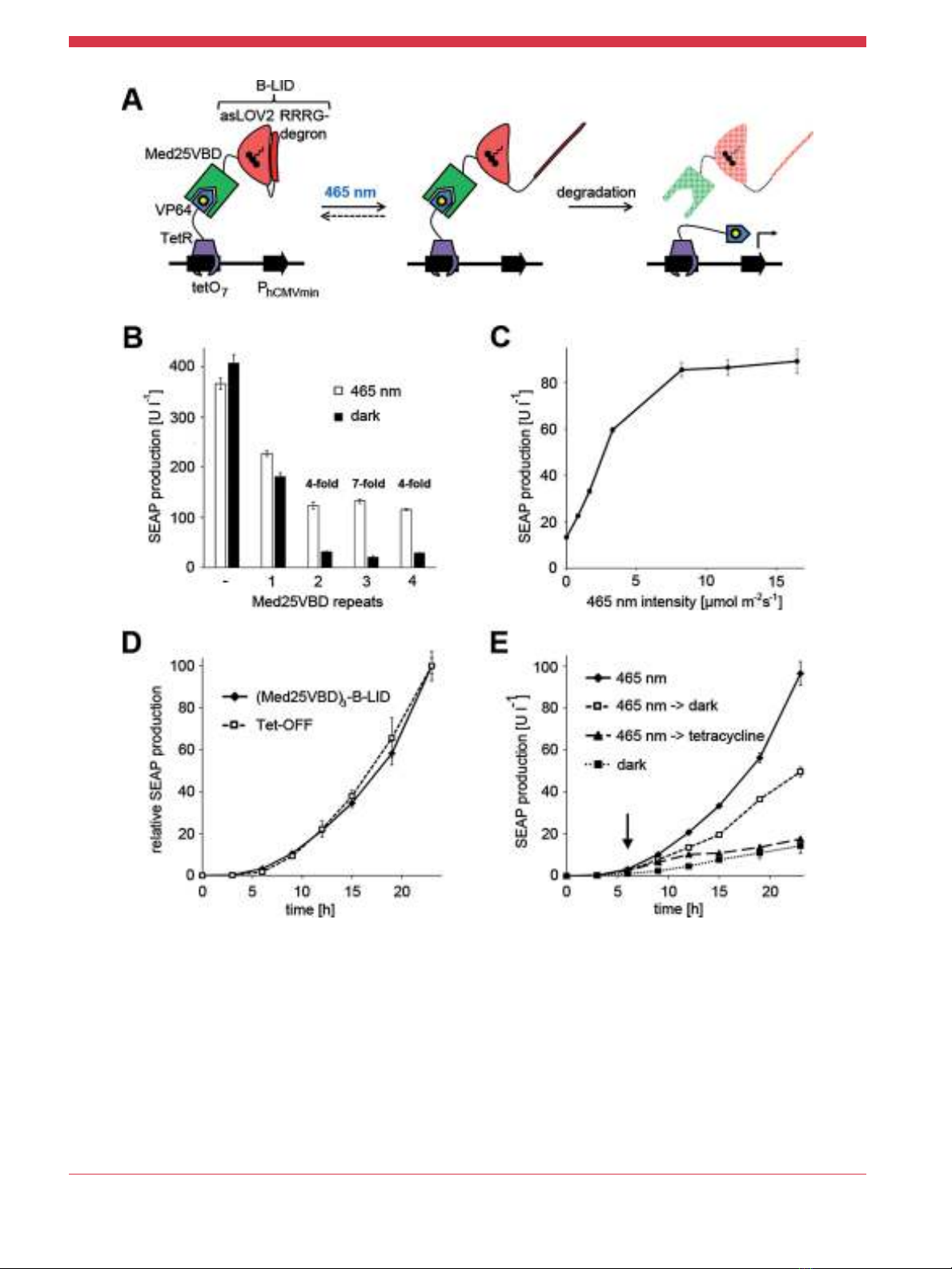
Figure 3.Light-regulated inhibition of the Tet-OFF system. A: Mode of function. In the dark, the VP64 transactivation domain is inhibited by Med25VBD that is fused to the B-LID
degradation tag, resulting in a shut-off of gene expression. Upon illumination with blue (465 nm) light, the Ja-helix of B-LID unravels, exposing the RRRG-degradation signal.
Consequently, Med25VBD is degraded and gene expression is initiated via VP64. B: Optimization of the blue light-responsive VP64-inhibitor. HEK-293T cells were transfected with the
Tet-OFF system (pKM551 and pMF111) alongside the light-regulated VP64-inhibitor with either one, two, three or four repeats of Med25VBD fused to B-LID. Control cells were
transfected with the Tet-OFF system and an empty plasmid backbone. All plasmids were transfected in equal amounts (w:w). Twenty-four hours post-transfection, the culture
medium was replaced with fresh medium and the cells were illuminated with 465 nm light (10 mmol m
2
s
1
) or incubated in the dark for 24 h prior to quantification of the SEAP
reporter. C: Dose-response curve. HEK-293T cells were transfected with the Tet-OFF system and the optimized light-responsive VP64-inhibitor (pKM546). Twenty-four hours post-
transfection the culture medium was replaced with fresh medium and the cells were illuminated for 24 h with 465 nm light of increasing intensities or incubated in the dark.
Afterwards, SEAP production was quantified in the culture medium. D: Comparison of the induction of gene expression from the basic Tet-OFF system and from the Tet-OFF system
with the optogenetic upgrade. HEK-293T cells were transfected with the Tet-OFF system (pKM551 and pMF111) or with the Tet-OFF system and the light-inducible inhibitor (pKM546)
and cultivated in the presence of tetracycline or in the dark, respectively. Twenty-four hours post transfection the cells were washed and illuminated with 465 nm light in the absence
of tetracycline. Reporter production was quantified at the indicated points in time. E: Kinetics of light-responsive gene expression. HEK-293T cells were transfected for SEAP
production from the Tet-OFF system with the optogenetic upgrade (pKM551, pMF111, and pKM546). Twenty-four hours post-transfection the culture medium was replaced with fresh
medium and the cells were illuminated with 465 nm light (10 mmol m
2
s
1
). Six hours later, tetracycline was added, or the cells were moved to the dark. Control cells were illuminated
with 465 nm light or incubated in the dark for the entire experiment. Reporter production was quantified at the indicated points in time and the addition of tetracycline or the switch to
darkness is indicated by an arrow. Data are means SD (B þC, n¼4; D þE, n¼3).
4Biotechnology and Bioengineering, Vol. 9999, No. xxx, 2015

neutral density filters (Schott) that were placed on top of the culture
dishes and the intensity was measured, using a quantum sensor
(LI-COR, prod. no. Q45045). All cell-handling involving the blue
light-inducible expression systems was done under safe 522 nm
light.
Reporter Gene Assays
The reporter SEAP was quantified in the cell culture medium, using
a colorimetric assay as described elsewhere (Muller et al., 2014c).
The numeric values for the data shown in Figures 1–3 are provided
in Supplementary Tables S4-S9.
We would like to thank Maria Karlsson for providing us with the plasmid
pLMK116.
References
Bacchus W, Lang M, El-Baba MD, Weber W, Stelling J, Fussenegger M. 2012.
Synthetic two-way communication between mammalian cells. Nat Biotechnol
30(10):991–996.
Beerli RR, Segal DJ, Dreier B, Barbas CF, III. 1998. Toward controlling gene
expression at will: specific regulation of the erbB-2/HER-2 promoter by using
polydactyl zinc finger proteins constructed from modular building blocks. Proc
Natl Acad Sci USA 95(25):14628–14633.
Bonger KM, Rakhit R, Payumo AY, Chen JK, Wandless TJ. 2014. General method for
regulating protein stability with light. ACS Chem Biol 9(1):111–115.
Fussenegger M, Morris RP, Fux C, Rimann M, von Stockar B, Thompson CJ, Bailey JE.
2000. Streptogramin-based gene regulation systems for mammalian cells. Nat
Biotechnol 18(11):1203–1208.
Gautier A, Gauron C, Volovitch M, Bensimon D, Jullien L, Vriz S. 2014. How to control
proteins with light in living systems. Nat Chem Biol 10(7):533–541.
Gitzinger M, Kemmer C, El-Baba MD, Weber W, Fussenegger M. 2009. Controlling
transgene expression in subcutaneous implants using a skin lotion containing
the apple metabolite phloretin. Proc Natl Acad Sci USA 106(26):10638–10643.
Gossen M, Bujard H. 1992. Tight control of gene expression in mammalian cells by
tetracycline-responsive promoters. Proc Natl Acad Sci USA 89(12):5547–5551.
Milbradt AG, Kulkarni M, Yi TF, Takeuchi K, Sun ZYJ, Luna RE, Selenko P, Naar AM,
Wagner G. 2011. Structure of the VP16 transactivator target in the mediator. Nat
Struct Mol Biol 18(4):410–U36.
Mitta B, Rimann M, Ehrengruber MU, Ehrbar M, Djonov V, Kelm J, Fussenegger M.
2002. Advanced modular self-inactivating lentiviral expression vectors for
multigene interventions in mammalian cells and in vivo transduction. Nucleic
Acids Res 30(21):e113.
Muller K, Engesser R, Schulz S, Steinberg T, Tomakidi P, Weber CC, Ulm R, Timmer J,
Zurbriggen MD, Weber W. 2013. Multi-chromatic control of mammalian gene
expression and signaling. Nucleic Acids Res 41(12):e124.
Muller K, Engesser R, Timmer J, Zurbriggen MD, Weber W. 2014a. Orthogonal
optogenetic triple-gene control in Mammalian cells. ACS Synth Biol 3(11):796–
801.
Muller K, Naumann S, Weber W, Zurbriggen MD. 2014b. Optogenetics for gene
expression in mammalian cells. Biol Chem. 396(2):145–52.
Muller K, Zurbriggen MD, Weber W. 2014c. Control of gene expression using a
red- and far-red light-responsive bi-stable toggle switch. Nat Protoc 9(3):
622–632.
Schonig K, Bujard H, Gossen M. 2010. The power of reversibility regulating gene
activities via tetracycline-controlled transcription. Methods Enzymol 477:429–
453.
Seipel K, Georgiev O, Schaffner W. 1992. Different activation domains stimulate
transcription from remote (’enhancer’) and proximal (’promoter’) positions.
EMBO J 11(13):4961–4968.
Urlinger S, Helbl V, Guthmann J, Pook E, Grimm S, Hillen W. 2000. The p65 domain
from NF-kappaB is an efficient human activator in the tetracycline-regulatable
gene expression system. Gene 247(1-2):103–110.
Weber W, Fussenegger M. 2011. Molecular diversity-the toolbox for synthetic gene
switches and networks. Curr Opin Chem Biol 15(3):414–420.
Weber W, Fux C, Daoud-el Baba M, Keller B, Weber CC, Kramer BP, Heinzen C, Aubel
D, Bailey JE, Fussenegger M. 2002. Macrolide-based transgene control in
mammalian cells and mice. Nat Biotechnol 20(9):901–7.
Weitzman M, Hahn KM. 2014. Optogenetic approaches to cell migration and beyond.
Curr Opin Cell Biol 30C:112–120.
Yamit-Hezi A, Dikstein R. 1998. TAFII105 mediates activation of anti-apoptotic
genes by NF-kappaB. EMBO J 17(17):5161–5169.
Yamit-Hezi A, Nir S, Wolstein O, Dikstein R. 2000. Interaction of TAFII105 with
selected p65/RelA dimers is associated with activation of subset of NF-kappa B
genes. J Biol Chem 275(24):18180–18187.
Yang FJ, DeBeaumont R, Zhou S, Naar AM. 2004. The activator-recruited cofactor/
Mediator coactivator subunit ARC92 is a functionally important target of
the VP16 transcriptional activator. Proc Natl Acad Sci USA 101(8):
2339–2344.
Supporting Information
Additional supporting information may be found in the online
version of this article at the publisher’s web-site.
M€
uller et al.: An Optogenetic Upgrade for the Tet-OFF 5
Biotechnology and Bioengineering 5



















![Bộ Thí Nghiệm Vi Điều Khiển: Nghiên Cứu và Ứng Dụng [A-Z]](https://cdn.tailieu.vn/images/document/thumbnail/2025/20250429/kexauxi8/135x160/10301767836127.jpg)
![Nghiên Cứu TikTok: Tác Động và Hành Vi Giới Trẻ [Mới Nhất]](https://cdn.tailieu.vn/images/document/thumbnail/2025/20250429/kexauxi8/135x160/24371767836128.jpg)





