
REVIEW ARTICLE
Structural recognition of DNA by
poly(ADP-ribose)polymerase-like zinc finger families
Stefania Petrucco and Riccardo Percudani
Department of Biochemistry and Molecular Biology, University of Parma, Italy
Introduction
PARP-like zinc fingers (zf-PARP) are zinc coordi-
nated protein domains that assist the DNA structure
recognition of different eukaryotic enzymes, and owe
their name to the proteins where these domains were
identified for the first time, namely poly(ADP-ribose-
polymerases (PARPs) [1,2]. Beyond PARPs, other
enzymes involved in the DNA metabolism are also
characterized by the presence of zf-PARPs and,
among them, mammalian DNA ligases III and plant
DNA 3¢phosphatases have been studied in some
detail.
PARPs
PARPs are a family of abundant eukaryotic enzymes
that catalyse the reversible, NAD
+
-dependent poly
ADP-ribosylation of protein substrates. PARP-1,
the most represented and studied member of the
PARP family, is characterized by the presence of two
unusually long zinc fingers (zf-PARPs), that are
positioned upstream of the catalytic domain (Fig. 1).
zf-PARPs mediate DNA recognition by PARP-1 and
were initially termed as nick-sensors due to their spe-
cific binding to nicked DNA [3]. It was subsequently
demonstrated that zf-PARPs also recognize other
DNA structures, including double-strand breaks, three-
and four-way junctions, hairpins, bubbles, etc. [4–8].
Importantly, zf-PARPs represent the regulatory
domain of PARP-1, and they are required for inducing
enzyme activity upon DNA recognition [8–10]. The
amount of activation depends upon the bound DNA
structure as well as upon the relative concentrations of
NAD
+
and of ATP [11,12]. PARP-1 is its own best
substrate; other substrates include histones, DNA
synthesis and repair enzymes, topoisomerases, tran-
scription factors, centromeric proteins, etc. [13–18].
Keywords
DNA binding; DNA damage; PARP;
phylogenesis; zinc fingers
Correspondence
S. Petrucco, Department of Biochemistry
and Molecular Biology, Univesrity of Parma,
Parco Area delle Scienze 23 ⁄A,
I-43100 Parma, Italy
Fax: +39 0521 905151
Tel: +39 0521 905149
E-mail: petrucco@unipr.it
(Received 14 November 2007, revised 17
December 2007, accepted 24 December
2007)
doi:10.1111/j.1742-4658.2008.06259.x
PARP-like zinc fingers (zf-PARPs) are protein domains apt to the recogni-
tion of multiple DNA secondary structures. They were initially described
as the DNA-binding, nick-sensor domains of poly(ADP-ribose)polymerases
(PARPs). It now appears that zf-PARPs are evolutionary conserved in the
eukaryotic lineage and associated with various enzymes implicated in
nucleic acid transactions. In the present study, we discuss the functional
and structural data of zf-PARPSs in the light of a comparative analysis of
the protein family. Sequence and structural analyses allow the definition of
the conserved features of the zf-PARP domain and the identification of five
distinct phylogenetic groups. Differences among the groups accumulate on
the putative DNA binding surface of the PARP zinc-finger fold. These
observations suggest that different zf-PARP types have distinctive recogni-
tion properties for DNA secondary structures. A comparison of various
functional studies confirms that the different finger types can accomplish a
selective recognition of DNA structures.
Abbreviations
FI, N-terminal finger of PARP; FII, second finger of PARP; G1–5, groups, 1 to 5; PARP, poly(ADP-ribose)polymerase; zf-PARP, PARP-like zinc
fingers.
FEBS Journal 275 (2008) 883–893 ª2008 FEBS. No claim to original Italian government works 883

The nick sensing activity of zf-PARPs has sustained
the general opinion that DNA breaks are the major
sites of PARP-1 modifying activity. In recent studies,
however, a new property of DNA recognition by
PARP-1 has been described, which stresses the more
general aptitude of PARP-1 to function as a chromatin
modifier [16,19]. According to Kim et al. [16], PARP-1
is specifically bound to nucleosomes of nontranscribed,
H1-histone-free chromatin domains. The authors pro-
pose that PARP-1 structures a silent, but ready-to-be-
open, chromatin conformation, where the activity of
nucleosome-bound PARP-1 (but not of unbound
PARP) is regulated by the relative concentrations of
NAD and ATP.
Mammalian DNA ligase III and plant DNA 3¢
phosphatases
Mammalian DNA ligases III and plant DNA 3¢phos-
phatases represent the other two types of enzymes for
which zf-PARPs have been described [20–23].
As shown in Fig. 1, DNA ligase III bears a single
zf-PARP, whereas Arabidopsis DNA 3¢phosphatase
has three such fingers. Both enzymes are implicated in
single-strand DNA repair processes, which are respon-
sible for removing damage that has occurred on either
DNA filament, thus anticipating the occurrence of
much more dangerous double-strand DNA breaks.
DNA ligase III is an ATP dependent DNA ligase
that appears to be a repair specific enzyme. The speci-
ficity might originate from its interactions within a
single-strand DNA repair complex, which restricts
enzymatic activities to the damaged DNA position
[20,21].
The distinctive feature of DNA ligase III with respect
to other DNA ligases is the presence of a zf-PARP at
its N-terminus. Similar to PARP-1 fingers, the ligase
finger also recognizes different DNA secondary struc-
tures, such as nicks and cruciforms. In the case of
ligase III, however, and in contrast to PARP-1, DNA
recognition by the finger does not have an obvious
influence on enzymatic activity [20,21]. Curiously, it
would even seem that, in the case of the ligase III, the
zf-finger domain competes with the catalytic domain
for nicked DNA binding. Leppard et al. [24] suggested
that the DNA ligase III finger recognizes and interacts
with single strand breaks, when DNA ligase III, and
possibly the associated single strand DNA repair
complex, is bound to negatively charged, auto modified
PARP-1. Furthermore, it has been suggested that DNA
ligase III finger stimulates rejoining of DNA strand
breaks at sites of clustered damage [21]. Yet, a clear role
for the DNA ligase finger in vivo has not emerged.
Plant DNA 3¢phosphatases are phosphoesterases
that can restore functional 3¢DNA ends by specifically
removing 3¢blocking phosphates. This is a necessary
step in DNA repair pathways because DNA polyme-
rases and DNA ligases can only process DNA that
carries free 3¢OH ends.
Plant, but not animal, 3¢DNA phosphatases, bear a
unique N-terminal region comprising multiple copies
of zf-PARPs. As for ligase III and PARP-1, fingers
can bind to different DNA secondary structures but,
similar to ligase III and in contrast to PARP-1, DNA
binding does not control the enzymatic activity of
plant DNA 3¢phosphatases [22,23].
zf-PARPs are thus components of a very abundant
chromatin modifier, PARP-1, and they are necessary
for binding at specific DNA sites. They are also found
in DNA repair enzymes that do not require zf-PARPs
to recognize DNA damages. DNA ligases and 3¢DNA
phosphatases can obviously bind damaged DNA
(nicked and 3¢blocked, respectively) via the active site
of their catalytic domains.
Here, we address questions and propose answers
concerning the functional differences existing between
zf-PARPs, which have been proposed to share binding
specificity in different protein contexts [21,25].
The zf-PARP family
Searches in the DNA database for zf-PARP sequences
immediately show that this protein module is not
unique to the three enzymes mentioned above. More
Fig. 1. Domain architecture of the charac-
terized zf-PARP proteins. Only zf-PARPs and
associated catalytic domains are indicated,
according to Pfam annotation: DNA_ligase,
DNA ligase III; PNK3P, polynucleotide
kinase 3 phosphatase. Proteins are drawn to
scale and the limits of each domain are indi-
cated by corresponding amino acid posi-
tions.
zf-PARP families S. Petrucco and R. Percudani
884 FEBS Journal 275 (2008) 883–893 ª2008 FEBS. No claim to original Italian government works

protein types, including predicted polypeptides
encoded in the genomes of lower eukaryotes, also dis-
play zf-PARPs (Table 1).
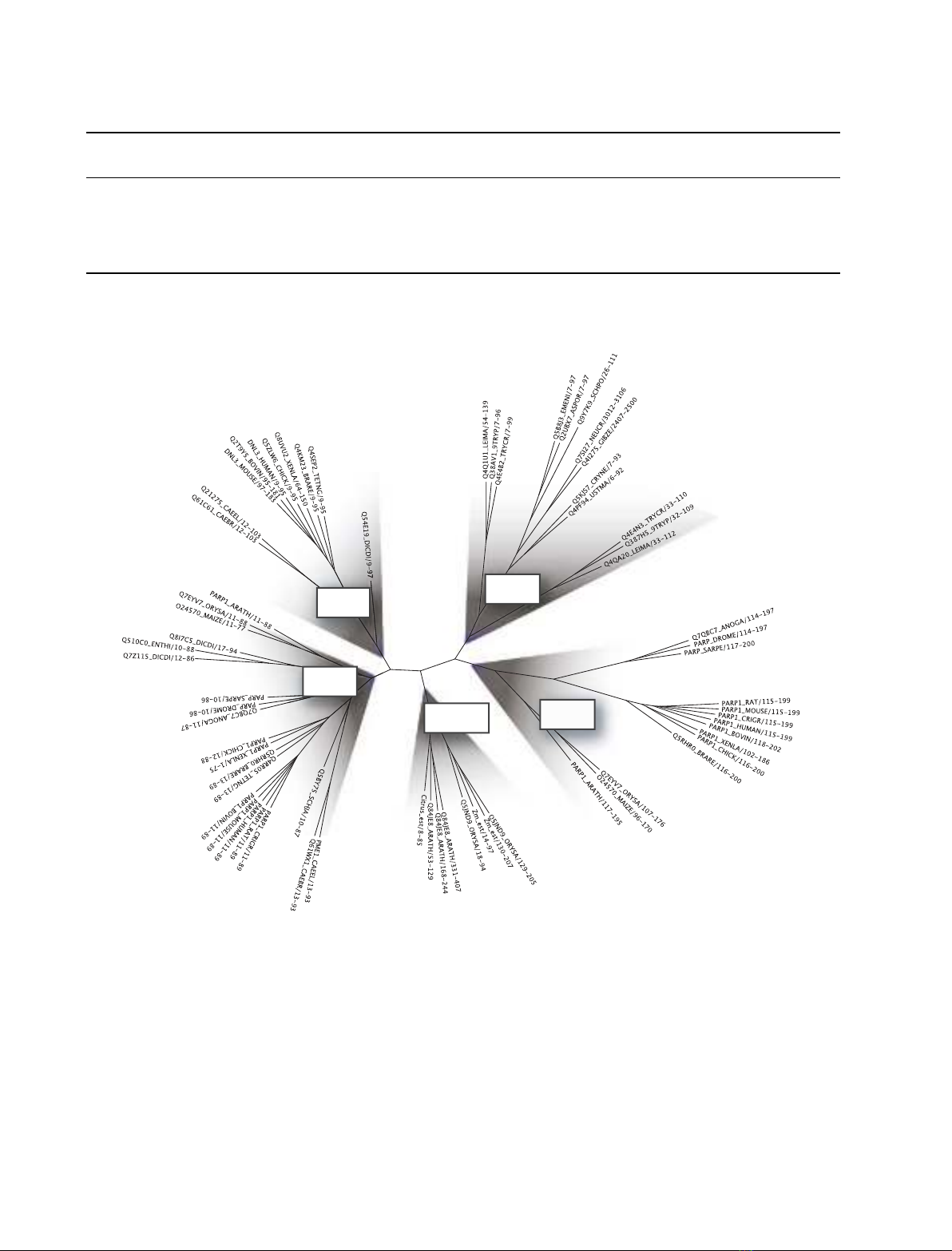
A sequence alignment of the complete family was
used to obtain the phylogenetic tree shown in Fig. 2,
which allows clustering zf-PARPs into five different
groups. Group one (G1) comprises the first, N-termi-
nal finger (FI) of PARPs. Group two (G2) comprises
DNA ligase III fingers and only includes animal and
mycetozoan sequences. Group three (G3) comprises
the zf-PARPs that are exclusively found in plant DNA
3¢phosphatases. Group four (G4) comprises the sec-
ond (FII) fingers of animal and plant PARPs. Interest-
ingly, all other eukaryotes appear to lack a second
finger in their PARPs. Group five (G5) comprises fun-
gal and protozoan sequences of putative DNA heli-
cases, high mobility group proteins and RNA binding
kinases. Furthermore, fingers of this group can be
orphan of any catalytic domain and simply be associ-
ated with low complexity protein regions. Given that
low complexity regions often provide sites for protein–
protein interactions, orphan fingers possibly provide
DNA binding functions to interacting protein com-
plexes.
Overall, this group composition suggests an ancient
origin for the zf-PARP module, which appears to be
generally associated with domains involved in nucleic
acid transactions. Because of the ancient separation of
all zf-PARP groups, the branching order of the deepest
nodes of the tree, and thus the relationships between
the different groups, cannot be assessed with high con-
fidence. In the case of PARP-1, however, it appears
that the acquisition of a second finger predates animal
and plant divergence. This would imply that FII fin-
gers have been subsequently lost in some lineages (e.g.
Caenorhabditis elegans). The presence of an FII finger
in complex organisms might reflect additional roles
acquired by PARP-1 as a chromatin modifier. It is
interesting to note that FI and FII fingers of PARP-1
are clearly divergent and, indeed, PARP FI appears to
be more related to DNA ligase III fingers than to
PARP FII. In keeping with the phylogenetic analysis,
it was previously shown that the DNA ligase III finger
is specifically recognized by PARP FI, but not FII,
anti-antibodies [20]. These observations may indicate
that FI and FII fingers are not redundant and supply
diverse functions in PARP-1 activity. By contrast, all
DNA 3¢phosphatase fingers cluster in G3 and show
relatively recent duplications, which occurred after the
monocots–dicots split. Also, the number of fingers
associated with these enzymes is quite variable (from
one in Citrus to three in Arabidopsis), thus suggesting
that these fingers have redundant roles in DNA 3¢
phosphatases. Finally, no member of the group five
has yet been characterized. Future work might elicit
insights regarding the specific functions of zf-PARPs
of this group, which is the most heterogeneous in
terms of protein domain architecture.
When the sequences of different groups are com-
pared, a number of aligned positions show strong
amino acid conservation, allowing the definition of a
general signature of the zf-PARP domain (Fig. 3).
Beyond residues for zinc coordination, four hydropho-
bic and four charged amino acid residues appear to be
almost invariant in all finger types (Fig. 3A,B). Indeed,
some of the invariant residues have been functionally
tested in vitro and in vivo and shown to be essential for
DNA binding [21,26]. However two regions, named
region V1 and V2 in Fig. 3, are highly variable among
zf-PARPs, both in length and sequence. A clear signa-
ture of the group is only observable in the case of
G2 and G4 fingers, but conserved features can also be
noticed in the variable regions of other groups
(Fig. 4).In particular, V1 in G1 fingers displays a
prevalence of hydrophobic amino acid residues sepa-
rated by a highly conserved aspartic residue. In the
same region, G2 fingers display a predominance of
hydrophobic and small amino acid residues, whereas
G4 fingers have a prevalence of charged amino acids.
V1 in G3 and G5 fingers shows poor conservation. V2
is mostly conserved within G2, 3 and 4 fingers, with a
large majority of charged residues in G2 and a highly
conserved RxELxF motif in G4. V2 is shorter in
G3 fingers and characterized by conserved proline resi-
dues. Functional divergence among proteins of G5
could also account for the sequence heterogeneity
observed within this group.
In summary, the alignment of zf-PARPs suggests
that, in a very much conserved backbone scaffold, two
variable regions might be in charge of providing spe-
cific properties to the different zf-PARP groups.
zf-PARP structure
Multiple studies provide direct evidence that isolated
zf-PARP domains can recapitulate the binding proper-
ties of full-length proteins. Thus, structural features of
zf-PARPs are the basis of the DNA recognition. The
recent addition of two zf-PARP structures deriving
from structural genomics initiatives, corresponding to
the FI (PDB 1v9x) and FII (PDB 2cs2) fingers of
PARP-1, allows comparison with the published struc-
ture of the ligase III finger [25]. As expected, a similar
overall organization can be recognized within these
three structures, which belong to the glucocorticoid
receptor-like (DNA binding domain) super family. A
S. Petrucco and R. Percudani zf-PARP families
FEBS Journal 275 (2008) 883–893 ª2008 FEBS. No claim to original Italian government works 885

Table 1. List of the zf-PARP containing proteins considered in the present study. Sequences containing the zf-Parp domain were retrieved
from the Pfam entry PF00645 (http://pfam.sanger.ac.uk) with the addition of Zea mays and Citrus clementina polynucleotide 3-phosphatases,
which where deduced from EST assemblies. Sequences less than 90% identical were retained in the final set and utilized for phylogenetic
analysis. Catalytic domains are indicated according to Pfam annotation: DNA_ligase, DNA ligase III; PNK3P, polynucleotide kinase 3 phospha-
tase; PI3_PI4_kinase, phosphatidylinositol 3- and 4-kinase; Helicase_C, helicase conserved C-terminal domain.
ID
a
Protein description Length
Catalytic
domains
b
Organism name Taxon
Phylogenetic
Group
a
Q8I7C5_DICDI NAD
+
ADP-ribosyltransferase-1B 804 PARP Dictyostelium discoideum Mycetozoa G1
Q5RHR0_BRARE Novel protein similar to
vertebrate ADP-
ribosyltransferase
1013 PARP Brachydanio rerio Metazoa G1 + G2
Q7QBC7_ANOGA ENSANGP00000014723 995 PARP Anopheles gambiae str.
PEST
Metazoa G1 + G2
Q510C0 ENTHI_ Poly(ADP-ribose) polymerase 845 PARP Entamoeba histolytica Enthamoebidae G1
Q7Z115_DICDI NAD
+
ADP-ribosyltransferase-1A 938 PARP Dictyostelium discoideum Mycetozoa G1
Q61WX1_CAEBR Hypothetical protein CBG04221 936 PARP Caenorhabditis briggsae Metazoa G1
PME1_CAEEL Poly(ADP-ribose)polymerase
pme-1
945 PARP Caenorhabditis elegans Metazoa G1
PARP_SARPE Poly(ADP-ribose)polymerase 996 PARP Sarcophaga peregrina Metazoa G1 + G2
PARP_DROME Poly(ADP-ribose)polymerase 994 PARP Drosophila melanogaster Metazoa G1 + G2
PARP1_XENLA Poly(ADP-ribose)polymerase 998 PARP Xenopus laevis Metazoa G1 + G2
PARP1_RAT Poly(ADP-ribose)polymerase 1 1014 PARP Rattus norvegicus Metazoa G1 + G2
PARP1_MOUSE Poly(ADP-ribose)polymerase 1 1013 PARP Mus musculus Metazoa G1 + G2
PARP1_HUMAN Poly(ADP-ribose)polymerase 1 1014 PARP Homo sapiens Metazoa G1 + G2
PARP1_CRIGR Poly(ADP-ribose)polymerase 1 1013 PARP Cricetulus griseus Metazoa G1 + G2
PARP1_CHICK Poly(ADP-ribose)polymerase 1 1011 PARP Gallus gallus Metazoa G1 + G2
PARP1_BOVIN Poly(ADP-ribose)polymerase 1 1016 PARP Bos taurus Metazoa G1 + G2
PARP1_ARATH Poly(ADP-ribose)polymerase 1 983 PARP Arabidopsis thaliana Viridiplantae G1 + G2
Q4KM23_BRARE Zgc:112973 752 DNA_ligase Brachydanio rerio Metazoa G4
Q5ZLW6_CHICK Hypothetical protein 902 DNA_ligase Gallus gallus Metazoa G4
Q8UVU2_XENLA DNA ligase III isoform alpha 988 DNA_ligase Xenopus laevis Metazoa G4
Q4SEP2_TETNG Chromosome undetermined
SCAF14615
873 DNA_ligase Tetraodon nigroviridis Metazoa G4
Q2T9Y5_BOVIN Similar to DNA ligase III 943 DNA_ligase Bos taurus Metazoa G4
DNL3_MOUSE DNA ligase 3 1015 DNA_ligase Mus musculus Metazoa G4
DNL3_HUMAN DNA ligase 3 922 DNA_ligase Homo sapiens Metazoa G4
Zm EST
b
EST assembly 462 PNK3P Zea mays Viridiplantae G3
Citrus EST
c
EST assembly 276 PNK3P Citrus clementina Viridiplantae G3
Q84JE8_ARATH Putative DNA nick-sensor
protein
694 PNK3P Arabidopsis thaliana Viridiplantae G3
Q5JND9_ORYSA Putative phosphoesterase 463 PNK3P Oryza sativa Viridiplantae G3
Q4I275_GIBZE Hypothetical protein 2729 PI3_PI4_kinase Gibberella zeae Fungi G5
Q7SI27_NEUCR Hypothetical protein
NCU00625.1
3409 PI3_PI4_kinase Neurospora crassa Fungi G5
Q4QA20_LEIMA DNA repair protein, putative 1092 Helicase_C Leishmania major Euglenozoa G5
Q387H5_9TRYP DNA repair protein, putative 984 Helicase_C Trypanosoma brucei Euglenozoa G5
Q4E4N3_TRYCR DNA repair protein, putative 983 Helicase_C Trypanosoma cruzi Euglenozoa G5
Q4RR05_TETNG Chromosome 14 SCAF15003 233 Tetraodon nigroviridis Metazoa G1
Q4Q1U1_LEIMA Hypothetical protein 285 Leishmania major Euglenozoa G5
Q21275_CAEEL Hypothetical protein 493 Caenorhabditis elegans Metazoa G5
Q5BY75_SCHJA SJCHGC03951 protein 165 Schistosoma japonicum Metazoa G1
Q54E19_DICDI SMAD ⁄FHA domain-containing
protein
895 Dictyostelium
discoideum AX4
Mycetozoa G5
Q61C61_CAEBR Hypothetical protein CBG13063 467 Caenorhabditis briggsae Metazoa G5
Q38AV1_9TRYP Hypothetical protein 240 Trypanosoma brucei Euglenozoa G5
Q4E4B2_TRYCR Hypothetical protein 230 Trypanosoma cruzi Euglenozoa G5
Q5KJS7_CRYNE Hypothetical protein 254 Cryptococcus
neoformans
Fungi G5
zf-PARP families S. Petrucco and R. Percudani
886 FEBS Journal 275 (2008) 883–893 ª2008 FEBS. No claim to original Italian government works
schematic view of the zf-PARP fold is shown in Fig. 5.
A three-stranded antiparallel bsheet characterizes the
N-terminal half of the domain (b1, b2 and b3in
Figs 3A and 5), with a long loop connecting b1 and b2
and containing two of the cysteine residues involved in
coordinating the zinc ion. The C-terminal half is
mainly ahelical (a1 and a2 in Figs 3A and 5),
with the third and the forth zinc-chelating residues
Table 1. (Continued).
ID
a
Protein description Length
Catalytic
domains
b
Organism name Taxon
Phylogenetic
Group
a
Q9Y7K9_SCHPO SPBC2A9.07c protein 274 Schizosaccharomyces
pombe
Fungi G5
Q4PF94_USTMA Hypothetical protein 546 Ustilago maydis Fungi G5
Q5B8J3_EMENI Hypothetical protein 279 Emericella nidulans Fungi G5
Q2UBX7_ASPOR Predicted protein 143 Aspergillus oryzae Fungi G5
a
Present study.
b
Deduced by the assembly of EST sequences DR813175, DN226524, DV517124, DR813176, DT649995.
c
Deduced by the
assembly of EST sequences DY292829, DY289144, DY300019.
G4 : PARP
Finger II
G5 : fungi
protozoa
G2 : DNA
Ligase
G1 : PARP
Finger I
G3 : PNK3P
Fingers I/II/III
Fig. 2. Phylogenetic relationships in the zf-PARP family. Alignment of the zf-Parp domains was carried out with the family Hidden Markov
model of Pfam using programs of the HMMER package [33]. Maximum-likelihood phylogeny was obtained with the PHYML program [34]. The
resulting unrooted maximum-likelihood tree was visualized with branch length adjustment for visibility enhancement using TREE ILLUSTRATOR.
Branches leading to the main phylogenetic groups are shadowed in gray and labelled according to the group composition. Sequenced are
indicated with identifiers (for details, see Table 1), followed by the sequence interval considered in the analysis.
S. Petrucco and R. Percudani zf-PARP families
FEBS Journal 275 (2008) 883–893 ª2008 FEBS. No claim to original Italian government works 887
















![Báo cáo seminar chuyên ngành Công nghệ hóa học và thực phẩm [Mới nhất]](https://cdn.tailieu.vn/images/document/thumbnail/2025/20250711/hienkelvinzoi@gmail.com/135x160/47051752458701.jpg)









