MINIREVIEW
Physico-chemical characterization and synthesis of neuronally active
a-conotoxins
Marion L. Loughnan and Paul F. Alewood
Institute for Molecular Bioscience, The University of Queensland, Brisbane, Australia
The high specificity of a-conotoxins for different neuronal
nicotinic acetylcholine receptors makes them important
probes for dissecting receptor subtype selectivity. New
sequences continue to expand the diversity and utility of
the pool of available a-conotoxins. Their identification
andcharacterizationdependonasuiteoftechniques
with increasing emphasis on mass spectrometry and micro-
scale chromatography, which have benefited from recent
advances in resolution and capability. Rigorous physico-
chemical analysis together with synthetic peptide chemistry
is a prerequisite for detailed conformational analysis and to
provide sufficient quantities of a-conotoxins for activity
assessment and structure–activity relationship studies.
Keywords:a-conotoxins; Conus; peptide synthesis; post-
translational modifications; sulfotyrosine.
Classification, primary structure and biology
of a-conotoxins
Cone snails are a group of hunting gastropods that
incapacitate their prey, which consists of worms, molluscs
or fish, by envenomation. Conotoxins from the venom of
cone snails are small disulfide-rich peptide toxins that act at
many voltage-gated and ligand-gated ion channels. They
can be grouped according to their molecular form into
several superfamilies, each defined by characteristic arrange-
ments of cysteine residues (not necessarily a single pattern),
and characteristic highly conserved precursor signal
sequence similarities. Individual conopeptide families
within a superfamily are denoted by Greek letters and
contain peptides that have a particular disulfide framework
and target homologous sites on a particular receptor [1].
Each of the characterized conopeptides is named using
a convention that indicates the activity (Greek letter), the
source species from which the peptide was first isolated
(Arabic letter(s)), the disulfide framework category (Roman
numeral) and the order of discovery within that category
(Arabic capital letter) [1]. For example a-AuIB belongs to
the a-conotoxin family and was the second peptide, B, with
that framework, I, isolated and reported from Conus aulicus
[1,2]. The names of some conotoxins deviate from this
nomenclature convention because their discovery preceded
its formulation. Hence some a-conotoxin names do not
conform to the alphabetical identifier system used to
indicate order of discovery of peptides with a specified
disulfide framework from the venom of any one species. The
framework identifiers I and II are both used in reference
to disulfide frameworks of the A superfamily without
distinction.
The A superfamily is so far comprised of the K
+
channel
blocking jA familiy and the aand aA families, which
together with the wfamily act at the nicotinic acetylcholine
receptor (nAChR). No aAorwconopeptides have been
reported to block neuronal nicotinic receptors with high
affinity. Rather, they are generally muscle-specific nicotinic
receptor antagonists [1]. The a-conotoxins fall into two
categories depending on whether they act at muscle-
type or neuronal-type receptors. The neuronally active
a-conotoxins are the focus of this minireview.
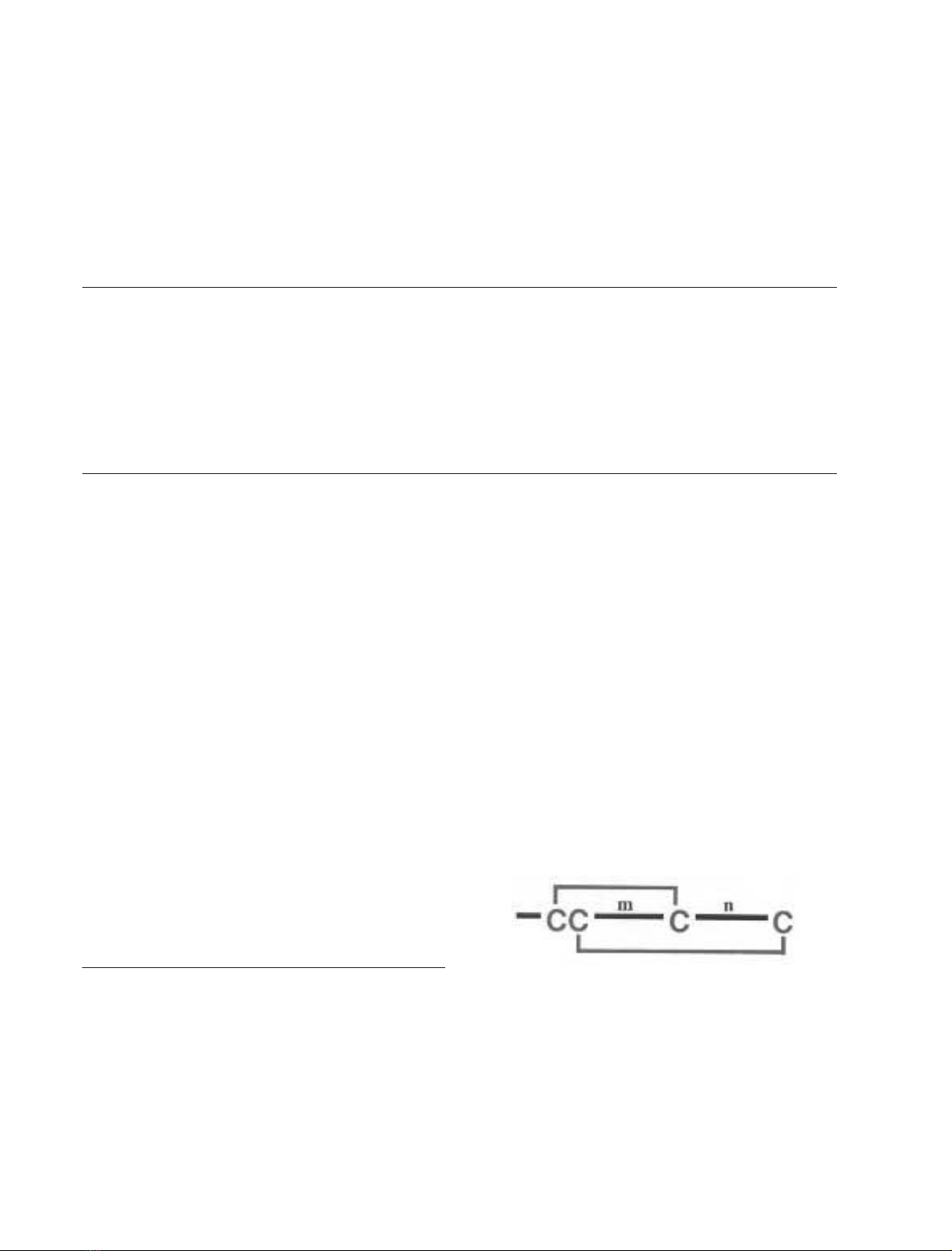
The known a-conotoxins consist of 12–19 amino acids.
Most a-conopeptides have four cysteine residues and the
general sequence GCCX
m
CX
n
C. The disulfide connectivity
is between alternate cysteine residues (I-III, II-IV).
The numbers of amino acid residues encompassed by the
second and third cysteine residues (m) and the third and
fourth cysteine residues (n) are the basis for a further
division into several structural subfamilies (a3/5, a4/3, a4/6
and a4/7) [1,3,4]. For example a4/6-AuIB belongs to the 4/6
disulfide loop size subgroup of the a-conotoxin family. The
neuronally active a-conotoxins are typically from the a4/7,
a4/6 and a4/3 subfamilies (Table 1). Peptides from the most
abundant a4/7subfamily are typically 16 residues in length
and range from 1600 to 1900 Da in mass. However
there have been recent additions to this subfamily in which
Correspondence to P. F. Alewood, Institute for Molecular Bioscience,
The University of Queensland, Brisbane, QLD 4072, Australia.
Fax: + 61 73346 2101, Tel.: + 61 73346 2982,
E-mail: P.Alewood@imb.uq.edu.au
Abbreviations:c-CRS, c-carboxylation recognition sequence;
nAChRs, nicotinic acetylcholine receptors; RT, retention time;
PTM, post-translational modification; TPST, tyrosyl-protein
sulfotransferase; TCEP, tris(2-carboxyethyl)phosphine; M-biotin,
maleimide-biotin; NEM, N-ethylmaleimide; IAM, iodoacetamide.
(Received 22 January 2004, revised 16 March 2004,
accepted 6 April 2004)
Eur. J. Biochem. 271, 2294–2304 (2004) FEBS 2004 doi:10.1111/j.1432-1033.2004.04146.x
there have been extensions at the N-terminus or C-terminus
to a length of up to 19 residues and the mass range has been
extended to almost 2200 Da [5]. For example, there are
three additional residues at the N-terminus in the case of
GID [5]. Peptides from the a4/3 and a4/6 subfamilies are
typically 12 and 15 residues, respectively. Unusually, EI has
the same disulfide framework as a4/7 conotoxins that target
neuronal nAChRs but has been reported to antagonize the
neuromuscular receptor as do the a3/5 and aA conotoxins
[1,6].
There is a conserved proline between the second and third
cysteine residues in almost all a-conotoxins except ImIIA
and ImII [7,8]. However, the former has not been confirmed
to be a neuronally active a-conotoxin, despite its sequence
similarity with ImI and ImII. There is also a conserved
serine residue between the second and third cysteine residues
in many a-conotoxins. The residue N-terminal to the first
cysteine residue of the sequence is in most cases glycine,
although exceptions are the recently isolated peptides GID
and PIA that have c-carboxyglutamic acid and proline,
respectively, in that position (Table 1) [5,9]. More generally,
the residues in the first loop tend to fit into defined
categories, whereas the second loop seems to have greater
heterogeneity of residues. There appears to be a relationship
between selected sequence motifs and receptor subtype
specificity and these sequence patterns may be a basis for
further defining subclasses within the neuronally active
members of the a-conotoxin family [9a].
There are many interesting features of the biology of
Conus species and the functional applications of the
a-conotoxins in their venom. It has been conjectured that
of the estimated 500 Conus species, each appears to make at
least one nAChR antagonist [1]. However, for some Conus
species pharmacological screening of crude venom samples
has not shown a-conotoxin activity (A. Nicke &
M. Loughnan, unpublished results). Nonetheless it has
become apparent that in any one species there may be
multiple peptides that target nAChRs [1], and it seems likely
that the complement of neuronally acting a-conotoxins in
one species may cover a range of subtype specificities. There
are examples of combinations of muscle-type and neuronal-
type a-conotoxins in a single species, particularly in the case
of the fish-eating Conus species. For example, C. geographus
venom contains the muscle-acting a-conotoxins GI and
GII together with the neuronally acting a-conotoxins
GIC and GID [1,5,10] and C. magus venom contains the
Table 1. Comparison of selected known a-conotoxins from a4/7, a4/6 and a4/3 families, their selectivity for mammalian nAChR subtypes, size, route of
discovery, method of synthesis and reference for synthesis. Asterisks (*) indicate an amidated C-terminus. The letters Oand Ydenote hydroxyproline
and sulfotyrosine, respectively. The letter Y
~~ denotes sulfotyrosine identified after original sequence was published. The symbol cdenotes
c-carboxyglutamic acid. Dashes indicate gaps in the sequence alignment. Mass (monoisotopic) in daltons, given for disulfide-bonded form.
Conserved cysteine residues are shown in red and a highly conserved proline in the first loop is shown in green. The peptides Vc1.1, GIC, ImII and
ImIIA were identified by prediction from the nucleic acid sequence. Other peptides were identified by isolation of the peptides from the venom ducts
in response to activity assays or by physico-chemical characteristics. Im peptides are from C. imperialis,AufromC. aulicus,MfromC. magus,Ep
from C. episcopatus,PnfromC. pennaceus,PfromC. purpurascens,GfromC. geographus and E from C. ermineus. Prey groups are denoted by p,
m, v for piscivores, molluscivores and vermivores, respectively. Discovery and synthesis methods were as follows: a, Discovered by peptide activity
at nAChRs in native tissues (e.g. bovine chromaffin cells, Aplysia neurons); b, Discovered by peptide activity at nAChRs heterologously expressed
in Xenopus oocytes; c, Discovered by peptide physico-chemical characteristics, and confirmed by synthesis and assay; d, Discovered by gene
sequencing with peptide sequence deduced from cDNA library obtained by RT-PCR of cone snail mRNA; e, Synthesised by Fmoc assembly,
trifluoroacetic acid cleavage and directed disulfide formation (off-resin); f, Synthesised by Fmoc assembly, modified trifluoroacetic acid cleavage
and air oxidation in ammonium bicarbonate for disulfide formation; g, Synthesised by tBoc assembly, HF cleavage and air oxidation in ammonium
bicarbonate for disulfide formation; N/A, not available.
Name Sequence Prey Selectivity Mass Methods Reference
ImI GCCSDPRCAWR----C*va7 1350.5 c,a,e [20]
ImIIA YCCHRGPCMVW----C*v N/A 1451.6 d [7]
ImII ACCSDRRCRWR----C*va7 1508.6 d,e [8]
AuIB GCCSYPPCFATNPD-C*ma3/b4 1571.6 b,e [2]
AuIA GCCSYPPCFATNSDYC*m (less active) 1724.6 b,e [2]
AuIC GCCSYPPCFATNSGYC*m (less active) 1666.6 b,e [2]
AnIA CCSHPACAANNQDYC*va3/b2, a7 1673.6 c,f [21]
AnIB GGCCSHPACAANNQDYC*va3/b2, a7 1787.6 a,f [21]
AnIC GGCCSHPACFASNPDYC*va3/b2, a7 1805.6 a,f [21]
MII GCCSNPVCHLEHSNLC*pa3b2; a6b2b3 1709.7 b,e [13]
EpI GCCSDPRCNMNNPDYC*ma3b2/a3b4; a7 1866.6 c,a,f [24]
Vc1.1 GCCSDPRCNYDHPEIC*ma3a7b4/a3a5b4 1805.7 d,a,g [22]
PnIA GCCSLPPCAANNPDY
~~
C*ma3b2
a
1701.6 a,g,e [58,51]
[A10L]PnIA GCCSLPPCALNNPDY
~~
C*a7
a
1663.7 g,e [60,51]
PnIB GCCSLPPCALSNPDY
~~
C*ma7
a
1716.6 a,g,e [59,51]
GIC GCCSHPACAGNNQHIC*pa3b2 1608.6 d,e [10]
GID IRDcCCSNPACRVNNOHVC#pa3b2 2184.6 b,g [5]
PIA RDPCCSNPVCTVHNPQIC*pa6b2b3 1980.8 d,e [9]
EI RDOCCYHPTCNMSNPQIC*p (muscle-type) 2091.8 a [6]
a
Synthesis method and activity refer to the unsulfated peptides.
FEBS 2004 Characterization and synthesis of a-conotoxins (Eur. J. Biochem. 271) 2295

muscle-acting a-conotoxin MI and the neuronally acting
a-conotoxin MII [11–13]. A biological interpretation of this
emerging pattern of paired ligand types is that prey capture
might rely on the combination of muscle-acting antagonists
to cause paralysis and neuronally acting antagonists to
inhibit the flight-or-fight response [10,14]. Although distinct
peptide complements have been attributed to individual
species [15], there are instances of a single a-conotoxin
sequence occurring in more than one species. For example,
GID from C. geographus has also been isolated from
C. tulipa venom [5].
Conus venoms together provide an array of ligands
with selectivity for various neuronal nAChR subtypes
(Table 1, [9a]). Evolutionarily, this diversity of toxins has
been generated by a hypermutation process that allows
protection of conserved cysteine residues and high substi-
tution rates for the intervening residues in the mature
toxin peptides [3,16,17]. Each venom peptide is processed
from a prepropeptide and the three defined regions of this
precursor (signal sequence, proregion, mature toxin) have
different rates of divergence [3,16]. Proposed diversifica-
tion mechanisms include gene duplication and subsequent
diversifying selection, or targeted gene mutation with
some sophisticated molecular regulation, perhaps based
on repair processes or recombination processes acting in
discrete exon regions [16–19]. Prey-driven diversifying
selection may be a factor for dominant expressed toxins,
given the feeding specificity of cone snail species [17]. It
has been suggested that nicotinic ligands from fish-hunters
are more likely than those from snail and worm-hunters
to target vertebrate nAChRs with high affinity [1]. Besides
the piscivores (fish-hunters) C. magus,C. geographus and
C. purpurascens, other species that have also yielded
neuronally active a-conotoxins include the vermivores
(worm-hunters) C. imperialis and C. anemone,andthe
molluscivores (mollusc-hunters) C. aulicus,C. victoriae,
C. pennaceus and C. episcopatus (Table 1) [2,5,7–10,13,20–
24]. Many more species are represented in a-conotoxin
sequence information contained in patent documents, for
example [25]. However these are not within the scope of
this review because their activities have not been reported,
although the patent applications reflect the commercial
interest in this class of conopeptides as potential candi-
dates for drug development.
Many of the neuronally active a-conotoxins show a
high conservation of the local backbone conformation
although the surfaces are unique [3,25a]. The common
structural scaffold suggests that the hypervariability of the
sidechain groups confers peptide specificity for different
neuronal nAChR subtypes [3]. Although the a-conotoxins
are considered to have rigid structures, multiple inter-
convertible isomers may exist in solution [1,3] (see also
below). This potential heterogeneity is an important issue
in the isolation, analysis and chemical synthesis of these
peptides.
Post-translational modifications of neuronally
active a-conotoxins
A feature of conotoxins in general is that they are relatively
richly endowed with a wide spectrum of post-translational
modifications (PTMs) and this aspect has been comprehen-
sively reviewed elsewhere [14,15,26]. The classical published
a-conotoxin sequences contained comparatively few mod-
ifications apart from disulfide bridges and C-terminal
amidation. However, more recently isolated peptides have
expanded the list of modifications and they now seem
comparable with the rest of the conotoxins in this respect
(Table 2). A thorough exploration of the significance of
these modifications for the function of a-conotoxins is yet to
be completed and reported.
Disulfide bridge formation is a basic feature of the
a-conotoxins with their defined cysteine spacing and
disulfide connectivity. Non-native disulfide-bonded cono-
peptides are often considered to be inactive but this is not
always the case. Intriguing results have been obtained in
structure-function studies of a synthetic variant of a-AuIB
with non-native disulfide bond connectivity, where an
enhancement of biological activity was observed [27].
Hydroxylation of proline has been observed in several
neuronally acting a-conotoxins but of these only GID has
been described [5]. Hydroxyproline also occurs elsewhere in
the A superfamily: in the muscle-specific a-EI, in aA-EIVA,
EIVB and PIVA and in w-PIIIE [1]. The significance of this
modification has not been determined. Many other cono-
toxins that contain multiple hydroxyproline residues also
have naturally occurring under-hydroxylated variants such
as in the example of TVIIA [28] and perhaps the modifi-
cation may not be critical. However there was no evidence
of a variant of GID with proline in place of the single
hydroxyproline residue [5].
Amidation of the C-terminus is a feature of most
conotoxins and so far occurs in the majority of the
a-conotoxins with the exceptions of GID and the muscle-
Table 2. Post-translational modifications (PTMs) in neuronal a-conotoxins.
PTMs in other conotoxins PTMS in a-conotoxins References
Disulfide bridge formation Yes all
Amidation of C-terminus Yes most (not in GID) [5]
Sulfation of tyrosine Yes EpI, PnIA, PnIB, AnIA, AnIB, AnIC [24,31,21]
Hydroxylation of proline Yes GID [5]
Carboxylation of glutamic acid Yes GID [5]
Cyclization of N-terminal Gln No
O-Glycosylation No
Bromination of tryptophan No
Isomerization of tryptophan No
Epimerization of other residues (L fiD) No
2296 M. L. Loughnan and P. F. Alewood (Eur. J. Biochem. 271)FEBS 2004
acting SII [1,5]. The functional significance of the nature of
the C-terminus in neuronal-type a-conotoxins has not been
established. However a recent study of the effects of an
amidated or free carboxyl C-terminus on activity of a
synthetic a-conotoxin (AnIB) at nAChR subtypes expressed
in oocytes showed subtype specific differences in activity [21].
Carboxylation of glutamic acid to c-carboxyglutamic
acid has been reported in GID [5]. It has been estimated that
10% of conopeptides contain c-carboxyglutamic acid and
the conantokins (NMDA receptor antagonists) have mul-
tiple c-carboxyglutamic acid residues that are important
for maintaining their three dimensional structure [15,29].
The existence of c-carboxylation recognition sequences
(c-CRSs) in conopeptide precursors from some Conus
species has been established and an enzyme responsible for
glutamic acid carboxylation in conantokin G has been
described [30]. The c-CRS regions in conantokin G and
bromosleeper peptides [30] are highly dissimilar suggesting
that thissequenceinformationcannotnecessarilybeextended
to other conopeptide families. c-CRSs in a-conotoxin
precursors have not been described.
Sulfation of tyrosine has been observed in EpI, PnIA,
PnIB, AnIA, AnIB and AnIC [21,24,31]. The mechanism of
sulfation of tyrosine by an enzyme, tyrosyl-protein sulfo-
transferase (TPST), has not been elucidated. It may involve
a recognition sequence in the peptide precursor [15] or a
consensus sequence in the mature peptide [32]. Alternat-
ively, secondary structure may be the major determinant of
sulfation and the TPST might broadly recognize any
sufficiently exposed tyrosine residue [33,34]. The sulfotyro-
sine-containing a-conotoxins have not previously been
reported to have substantially different activity from the
unmodified variants [15,24]. Recent comparisons of EpI
with [Y15]EpI, and AnIB with [Y16]AnIB found about
three-fold and ten-fold reduced activities, respectively, of the
unsulfated forms relative to the sulfated peptides [21,35]. In
the case of AnIB, tyrosine sulfation selectively influenced the
binding to the mammalian a7 but not the a3b2 subtype [21].
The effects of substitution of phosphotyrosine for sulpho-
tyrosine in a-conotoxins have not been investigated.
Post-translational modifications such as O-glycosylation
of serine or threonine residues, bromination of tryptophan,
isomerization of tryptophan or L fiD epimerization of
other residues have been observed in conotoxins from other
families [1,15] but not reported for a-conotoxins. However,
characterizations of a-conotoxins have not routinely inclu-
ded tests for isomerization and epimerization modifications.
Aspects of the biosynthesis of conotoxins in the cone snail
and the mechanisms for incorporation of PTMs have
garnered considerable interest [15]. Hypermutation of
amino acid residues is a feature of the mature toxins but
in contrast the prepropeptide precursor sequence, partic-
ularly the signal sequence, seems to be highly conserved for
each family of conotoxins [3,16]. Precursor sequences are
available for conotoxins from most families but there have
been relatively few precursors published in journals for the
a-conotoxins (Table 3). Nevertheless it is interesting to
examine the available prepropeptide sequence information
for ImIIA and Vc1.1, ImII, GIC and PIA, each of which
was identified by prediction from a genomic DNA clone
[7–10,22]. Comparison of the peptide sequences GID, EI,
ImIIA, GIC and PIA suggests that there are anomalies in
Table 3. Precursor sequence or cleavage site information for selected a-conotoxins. The mature peptide is underlined. The putative cleavage site for generation of the mature toxin from the prepropeptide at
one or more basic resisues is shown in bold.
Name Species Reference Sequence
GIC precursor C. geographus [10] SD GRNDAA--KA FDLI-SSTV- KKGCCSHPAC AGNNQHICGR RR
Vc1.1 precursor C. victoriae [22] MGMRMMFTVF LLVVLATTVV SSTSGRREFR GRNAAA--KA SDLV-SLTDK KRGCCSDPRC NYDHPEICG
ImIIA precursor C. imperialis [7] MGMRMMFTVF LLVVLATAVL PVTL-DRASD GRNAAANAKT PRLI-APFI- RDYCCHRGPC MVW----CG
ImII cleavage site C. imperialis [8]
a
RRACCSDRRC RWR----CG
ImI cleavage site C. imperialis [8]
a
RRGCCSDPRC AWR----CG
MII precursor C. magus [25] MFTVF LLVVLATTVV SFPS-DRASD GRNAAANDKA SDVITAL--- -KGCCSNPVC HLEHSNLCGR RR
AuIB precursor C. aulicus [25] MFTVF LLVVLATTVV SFTS-DRASD GRKDAA---- SGLI-ALTM- -KGCCSYPPC FATNPD-CGR RR
AuIA precursor C. aulicus [25] MFTVF LLVVLATTVV SFTS-DRASD GRKDAA---- SGLI-ALTI- -KGCCSYPPC FATNSDYCG
EpI precursor C. episcopatus [25] MFTVF LLVVLATTVV SFTS-DRASD SRKDAA---- SGLI-ALTI- -KGCCSDPRC NMNNPDYCG
PIA precursor C. purpurascens [9] SD GRDAAANDKA TDLI-ALTARRDPCCSNPVC TVHNPQICGR R
a
EMBL/Genbank/DDBJ databases accession numbers P50983, Q816R5.
FEBS 2004 Characterization and synthesis of a-conotoxins (Eur. J. Biochem. 271) 2297

relation to GID, EI and PIA, which each have an N-
terminal sequence motif RDX. It is tempting to speculate
about the relationship of this motif to the potential dibasic
cleavage sites for generation of the mature peptides from the
prepropeptides, particularly in those cases where the residue
at position X becomes post-translationally modified in the
mature peptide. However there are also many other
sequences reported in patent documents that have not been
considered here and for many of those the deduced cleavage
site does not have a dibasic motif [25]. There has been
increasing utility of molecular biology techniques for
identification of new a-conotoxin sequences. Reliance on
the gene sequence of a conopeptide alone without verifica-
tion from the peptide sequence might miss PTM sites, or
misidentify the cleavage site for generation of the mature
peptide. Much further work is necessary to elucidate the
mechanisms of incorporation of PTMs in the biosynthesis
of conopeptides. Appropriate methods of analysis need
to be addressed to ensure recognition of these PTMs,
if present, in the course of characterization of native
a-conotoxins.
Analysis of neuronally active a-conotoxins
using HPLC and MS, including identification
of post-translational modifications
Isolation and identification
Standard procedures for identification and isolation of
a-conotoxins generally incorporate separations using
reversed-phase HPLC, size exclusion or ion exchange
chromatography in combination with mass-based screening
and functional screening. Most of the a-conotoxins identi-
fied so far are relatively hydrophilic and hence tractable.
Mass-based screening entails searching by LC/MS or MS
alone for components with a mass in the defined range for
a-conotoxins and two disulfide bonds (identified by partial
reduction and alkylation studies and MS/MS). It may also
include diagnostic LC/MS for recognition of some post-
translational modifications and possibly MS/MS for recog-
nition of conserved sequence motifs. The small size range
of a-conotoxins would seem ideal for MS-based sequence
determination. However, complete de novo sequencing of
conopeptides by MS/MS is still considered experimental,
and primary sequence information is usually obtained by
Edman degradation sequencing, interpreted in conjunction
with MS data for the intact molecule. Efficient sequence
analysis usually requires that the peptides are reduced and
the cysteine residues alkylated in order to verify their
identification. Although there are standard procedures for
reduction and alkylation, their application to conopeptide
analysis and characterization is by no means trivial, and
often optimization on a case-by-case basis is required [14].
Sample amounts may be limiting even with the enhanced
sensitivity of current automated sequence analysis instru-
ments.
Chromatography and structural heterogeneity
Several a-conotoxins have a characteristic asymmetric peak
under standard reversed-phase HPLC elution conditions
[5,36]. The anomalous chromatographic behaviour is seen
particularly with a-CnIA, a-MI, a-GI and a-GID for both
native and synthetic forms and persists even after repeated
refractionation [5,36,37]. The asymmetry may be more
pronounced under isocratic elution conditions. It presum-
ably reflects structural heterogeneity and can be interpreted
as a slow interconversion between two conformers; this
conclusion has been supported by the results of structure
studies. NMR studies have shown the existence of two
distinct interconvertible conformers for GI and multiple
conformers of CnIA [36,37]. This heterogeneity may yet
prove to be an important feature of some a-conotoxins in
the understanding of structure-function relationships and
the interaction of these ligands with the nAChR.
Identification of PTMs
Methods for the characterization of PTMs in conotoxins
have been reviewed elsewhere, with particular emphasis
on the utility of MS [14], but specific aspects relevant to
a-conotoxins (identification of C-terminal amidation,
sulfotyrosine, hydroxyproline and c-carboxyglutamic acid)
are revisited here. The identification of the PTM is usually
confirmed by synthesis of the modified a-conotoxin and
comparison with the natural peptide.
Identification of the nature of the C-terminus. The
identification of C-terminal amidation or a free carboxyl
terminus in an a-conotoxin is usually straightforward with
the one mass unit difference between the two forms readily
apparent from the monoisotopic mass determined by high
resolution mass spectrometry. There may be difficulties in
interpretation of MS data when there are ambiguities
arising from, for example, asparagine to aspartic acid, or
glutamine to glutamic acid changes [14]. The a-conotoxins
EpI, PnIA, GIC, GID, AnIA and AnIB contain pairs of
asparagine residues [5,10,21,23,24] (Table 1), and deamida-
tion may confound MS data for these peptides.
Determination of sulfotyrosine. Identification of sulfo-
tyrosine in peptides is usually by mass spectrometry and
the lability of the sulfogroup in mass spectrometry
analysis allows recognition of the modification, and
differentiation of sulfotyrosine and phosphotyrosine [38].
Characterization of the sulfotyrosine-containing a-cono-
toxin EpI was undertaken by a combination of mass
spectrometry and modified amino acid analysis [24]. The
conotoxins a-PnIA and a-PnIB from C. pennaceus were
initially identified and reported as unmodified sequences
although an unidentified mass discrepancy had been
recognized [23]. The verification of tyrosine sulfation in
a-PnIA and a-PnIB and revision of those sequences were
made in an investigation of labile sulfo- and phospho-
peptides by electrospray MALDI and atmospheric
pressure MALDI mass spectrometry [31]. The sulfation
of three conotoxins from C. anemone was identified on
the basis of LC/MS under different conditions together
with the difference between the observed mass and that
predicted from primary Edman sequence data [21].
The presence of either sulfation or phosphorylation may
be indicated when liquid chromatography/electrospray
ionization mass spectrometry shows doubly protonated
species of the modified a-conotoxins with additional related
2298 M. L. Loughnan and P. F. Alewood (Eur. J. Biochem. 271)FEBS 2004


























