
Nucleosome positioning in relation to nucleosome spacing
and DNA sequence-specific binding of a protein
Rama-Haritha Pusarla*, Vinesh Vinayachandran* and Purnima Bhargava
Centre for Cellular & Molecular Biology, Hyderabad, India
Multifold compaction of DNA due to the presence of
nucleosomes on natural templates of eukaryotic RNA
polymerases results in transcriptional repression. Sev-
eral studies have established that histones and nucleo-
somes play an active role in regulating gene expression
in eukaryotic cells [1,2]. Gene-specific, localized config-
urations of in vivo chromatin in various genome
regions are found due to precise positioning of nucleo-
somes over the underlying DNA stretches [3,4]. Both
trans-acting factors and DNA sequences can determine
where histones will occupy the bound DNA. A nucleo-
some is positioned translationally when its histone–
DNA contacts are restricted to an identifiable stretch
of DNA, giving clear boundary zones. Further preci-
sion of the positioning can be achieved by restricting
the rotation of DNA over the histone octamer surface
(rotational setting), resulting in a defined phase ⁄orien-
tation of a particular base pair with respect to
histones. Nucleosomes on certain constitutively active
genes can be excluded due to rapid and tight associ-
ation of trans-acting factors with promoter elements
during the replication-coupled assembly of chromatin
in vivo [5]. On other genes, they are removed or
reshuffled through several chromatin-remodeling and
Keywords
chromatin assembly; ionic strength;
nucleosome positioning; nucleosome
spacing; protein boundary
Correspondence
P. Bhargava, Centre for Cellular & Molecular
Biology, Uppal Road, Hyderabad-500007,
India
Fax: +91 40 27160591
Tel: +91 40 27192603
E-mail: purnima@ccmb.res.in
*These authors contributed equally to this
work
(Received 12 October 2006, revised 2
March 2007, accepted 7 March 2007)
doi:10.1111/j.1742-4658.2007.05775.x
Nucleosome positioning is an important mechanism for the regulation of
eukaryotic gene expression. Folding of the chromatin fiber can influence
nucleosome positioning, whereas similar electrostatic mechanisms govern
the nucleosome repeat length and chromatin fiber folding in vitro. The
position of the nucleosomes is directed either by the DNA sequence or by
the boundaries created due to the binding of certain trans-acting factors to
their target sites in the DNA. Increasing ionic strength results in an
increase in nucleosome spacing on the chromatin assembled by the S-190
extract of Drosophila embryos. In this study, a mutant lac repressor protein
R3 was used to find the mechanisms of nucleosome positioning on a plas-
mid with three R3-binding sites. With increasing ionic strength in the pres-
ence of R3, the number of positioned nucleosomes in the chromatin
decreased, whereas the internucleosomal spacings of the positioned
nucleosomes in a single register did not change. The number of the posi-
tioned nucleosomes in the chromatin assembled in vitro over different plas-
mid DNAs with 1–3 lac operators changed with the relative position and
number of the R3-binding sites. We found that in the presence of R3,
nucleosomes were positioned in the salt gradient method of the chromatin
assembly, even in the absence of a nucleosome-positioning sequence. Our
results show that nucleosome-positioning mechanisms are dominant, as the
nucleosomes can be positioned even in the absence of regular spacing
mechanisms. The protein-generated boundaries are more effective when
more than one binding site is present with a minimum distance of
165 bp, greater than the nucleosome core DNA length, between them.
Abbreviations
IEL, indirect end-labeling; IPTG, isopropyl thio-b-D-galactoside; MNase, micrococcal nuclease; NRL, nucleosome repeat length.
2396 FEBS Journal 274 (2007) 2396–2410 ª2007 The Authors Journal compilation ª2007 FEBS

chromatin-modifying mechanisms. Thus, cells use
nucleosome positioning as a mechanism to include or
exclude the binding sites of trans-acting factors from
accessible chromatin regions by passively restricting
the position of nucleosomes therein [6,7]. It is now well
established that a nucleosome is not necessarily repres-
sive; rather, it can facilitate the activation of genes.
Specific positioning of the nucleosomes allows the
transcriptional machinery to work effectively in a chro-
matin environment. Folding of DNA by the histones
and positioned nucleosomes can bring two widely sep-
arated regulatory elements into juxtaposition in space
[8–10] or even orient the bound factors for productive
interactions.
Nucleosomal repeat length (NRL) is characteristic
for a species, suggesting that it is a regulated feature
of the chromatin [11]. Uniform spacing of nucleosomes
is proposed to promote the higher-order folding of
chromatin [12]. Chromatin folding, in turn, is reported
to influence nucleosome positioning [13]. The presence
of positioned nucleosomes at defined and limited loca-
tions may result in disruption of the uniformity in spa-
cing. However, it is not known whether nucleosome
spacing influences nucleosome positioning. The
observed longer repeat lengths on inactive genes as
compared to those observed on transcribed sequences
[14,15] suggest that the folding of the 10 nm chromatin
with beads on a string into a higher-order structure
requires a minimum spacing to be maintained between
core particles. Thus, nucleosome spacing and position-
ing appear to be correlated.
Ionic strength is reported to influence nucleosome
conformations [16,17] as well as their spacings [18].
Within an array of positioned nucleosomes, the ionic
strength effect dominates the sequence effect [19]. It
also influences chromatin folding, presumably by
modulating H1 association as well as interparticle
interactions [20,21]. Of the two methods of chromatin
assembly in vitro [22], the salt gradient dialysis method
deposits nucleosomes in a random fashion, and has
been useful for checking the ability of various DNA
sequences to position nucleosomes in vitro.ADro-
sophila embryonic extract, in contrast [23], can deposit
nucleosomes with regular spacing in a sequence-inde-
pendent manner in the presence of ATP. The in vitro
chromatin assembly carried out by cellular ⁄nuclear
extracts, giving uniformly spaced nucleosomes, is affec-
ted by parameters such as ionic strength, concentration
of linker histones, protein phosphorylation, and the
presence of core histone tails [18,24]. Spacing of nucle-
osomes is also influenced by DNA topology or histone
variants [19,25]. Using this system, binding of a
mutant lac repressor R3 (a small sequence-specific
DNA-binding prokaryotic protein) to its two sites,
183 bp apart, was shown to result in at least five trans-
lationally positioned nucleosomes in a single register
on a plasmid DNA [26]. In general, boundaries gener-
ated by proteins binding to the DNA restrict the
randomization of nucleosome positions [27]. It was
predicted that the nucleosomes close to a boundary
would be precisely positioned, whereas this precision
would decrease with distance from the boundary [28].
We have analyzed the effect of changing the ionic
strength and the number and spacing of the binding
sites on the protein-generated boundary for nucleo-
some positioning in both assembly systems. We have
found that the number of positioned nucleosomes in a
single register changes with the ionic strength of the
medium, although the spacing between them does not
change. The range of positioning effects of DNA
sequence-specific binding of a protein to chromatin
depends on the number of sites and the distance
between them.
Results
All the chromatin assemblies were constructed using
rat liver core histones and plasmid DNAs schematical-
ly depicted in the Fig. 1, in the presence or absence of
the R3 protein.
Number of positioned nucleosomes changes
with ionic strength
R3 is a mutant lac repressor protein that binds the lac
operator as a dimer with the same affinity as that of
the wild-type lac repressor but fails to tetramerize [29].
Binding of R3 protein to a plasmid DNA pU6LNS
(two lac operators at a distance of 183 bp) was reported
A
B
Fig. 1. Relative positions of the three lac operator sites in the plas-
mid DNAs. (A) Diagrammatic representation of the plasmid con-
structs with different numbers of lac operators. The solid
rectangular boxes and L1, L2 and L3 denote the first, second and
third lac operator sites. The distance in bp between each site is
given. (B) Schematic map of plasmid d35 with the shortest dis-
tance between L1 and L2.
R.-H. Pusarla et al. Mechanisms of nucleosome positioning
FEBS Journal 274 (2007) 2396–2410 ª2007 The Authors Journal compilation ª2007 FEBS 2397
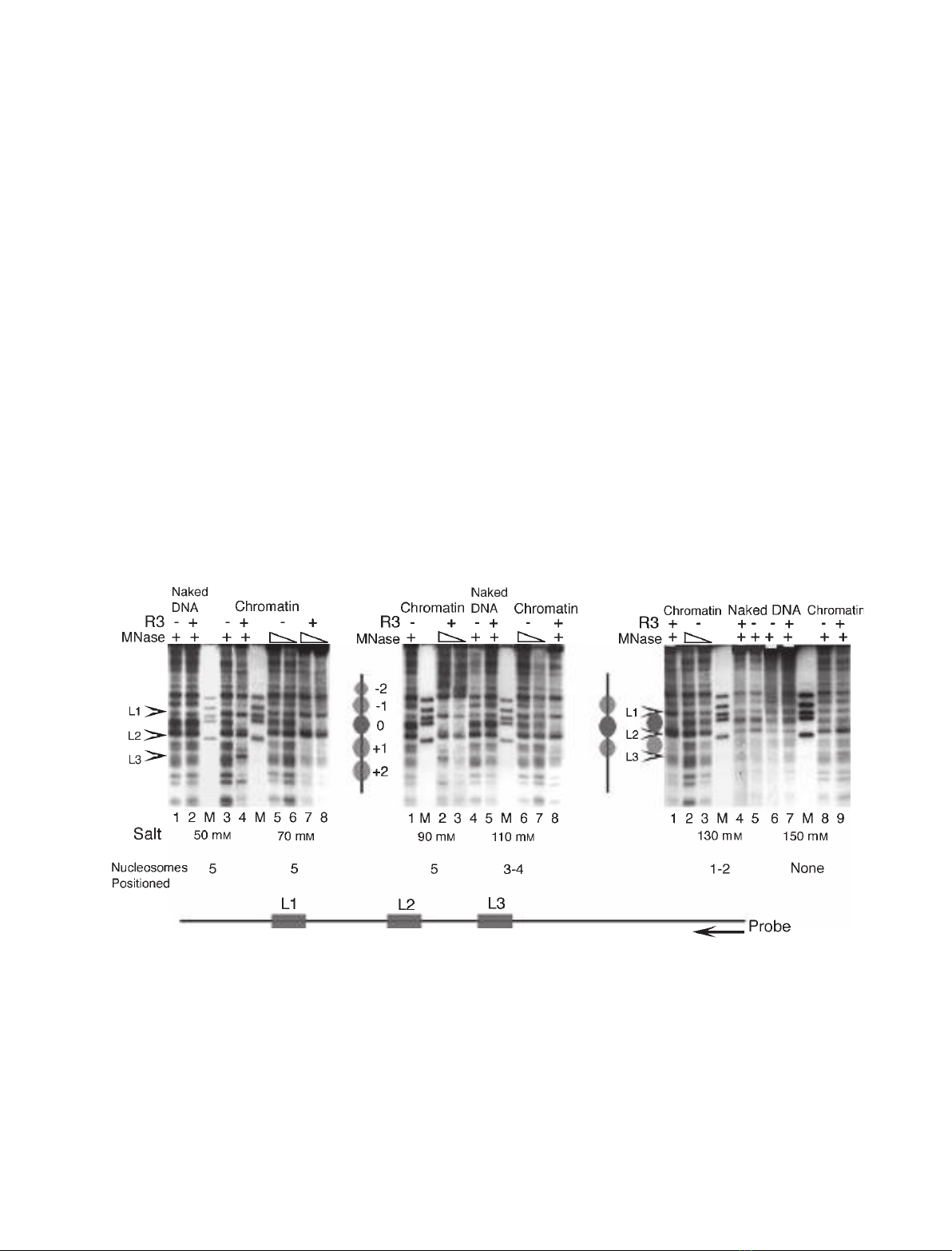
to result in the positioning of an array of five nucleo-
somes in a single register [26]. As R3 can bind to the
naked DNA at salt concentrations as high as 0.4 m
(not shown), we looked at the effect of increasing ionic
strength on nucleosome positioning due to R3 binding
on the plasmid pU6lac3 by using the indirect end-
labeling (IEL) method of chromatin structure analysis
(Fig. 2). The micrococcal nuclease (MNase) digestion
pattern of the naked DNA in the IEL analysis
(Fig. 2A, lane 1; Fig. 2B, lane 4; Fig. 2C, lanes 5 and
6) did not change with the binding of R3 (Fig. 2A,
lane 2; Fig. 2B, lane 5; Fig. 2C, lanes 4 and 7), as
small footprints could not be resolved in the agarose
gels. The digestion pattern did not change even with
the deposition of histones (Fig. 2A, lanes 3, 5 and 6;
Fig. 2B, lanes 1, 6 and 7; Fig. 2C, lanes 2, 3 and 8) at
every ionic strength, suggesting there are no preferred
locations for nucleosome assembly on the plasmid.
However, in the presence of R3, in a single register,
five positioned nucleosomes at a salt concentration of
50–90 mm(Fig. 2A, lanes 4, 7 and 8; Fig. 2B, lanes 2
and 3) and three positioned nucleosomes at a salt con-
centration of 110 mm(Fig. 2B, lane 8) could be seen.
In comparison to this, two positioned nucleosomes
could be seen even at a salt concentration of 130 mm,
whereas none were seen at a salt concenration of
150 mm(Fig. 2C, lanes 1 and 9). These results show
that with increasing ionic strength, fewer positioned
nucleosomes are aligned in a single register.
As positioned nucleosomes are seen in Fig. 2 only in
the presence of R3, we used DNaseI footprinting to
confirm the specific binding of R3 at L1 and L2 at all
the salt concentrations. As no positioned nucleosomes
were seen at a monovalent salt level of 150 mmin the
IEL analysis of Fig. 2C, no protection was seen
between L1 and L2 on the DNA subjected to chroma-
tin assembly in the representative gel at this ionic
strength (Fig. 3A). Chromatin assembly in our system,
as judged by the generation of MNase-resistant nucleo-
somal ladders, was found to be normal up to 110 mm
salt, whereas only a few nucleosomal bands could be
seen at higher salt levels (not shown). The binding of
R3 did not disrupt the nucleosomal ladders close to
the three distinct sites located at different distances
from each other (Fig. 3B). Therefore, the loss of posi-
tioned nucleosomes at higher ionic strengths is not due
AB C
Fig. 2. Nucleosome positioning in the presence of R3 at different ionic strengths. IEL analysis of the chromatin structure of plasmid pU6lac3
assembled with S-190 extract in the absence or presence of R3 at various salt concentrations. Nucleosome positions are numbered and
marked with ellipses, and arrowheads indicate the positions of lac operators, marked L1–L3. The total number of the positioned nucleo-
somes in a single register is given under each salt level. The 5¢-end of the radiolabeled oligonucleotide probe (arrowhead) hybridized 709 bp
downstream of the third lac operator, L3, as shown at the bottom of the figure. (A) Structure analysis of the chromatin assembled at 50 or
70 mMsalt. Lanes 1 and 2 show naked DNA digestion patterns, and lanes 3–8 represent chromatin. R3 is absent in lanes 1, 3, 5 and 6,
whereas chromatin was assembled at 50 mMsalt for lanes 1–4, and at 70 mMsalt for lanes 5–8. (B) IEL analysis for the chromatin assem-
bled at 90 and 110 mMsalt concentrations. Naked DNA (lanes 4 and 5) and chromatin assembled in the absence or presence of R3 protein
are shown. R3 was added to lanes 2, 3, 5 and 8. (C) IEL analysis for the chromatin assembled at 130 and 150 mMsalt. Naked DNA
(lanes 4–7) and chromatin assembled in the absence or presence of R3 protein are shown. R3 was added to lanes 1, 4, 7 and 9.
Mechanisms of nucleosome positioning R.-H. Pusarla et al.
2398 FEBS Journal 274 (2007) 2396–2410 ª2007 The Authors Journal compilation ª2007 FEBS
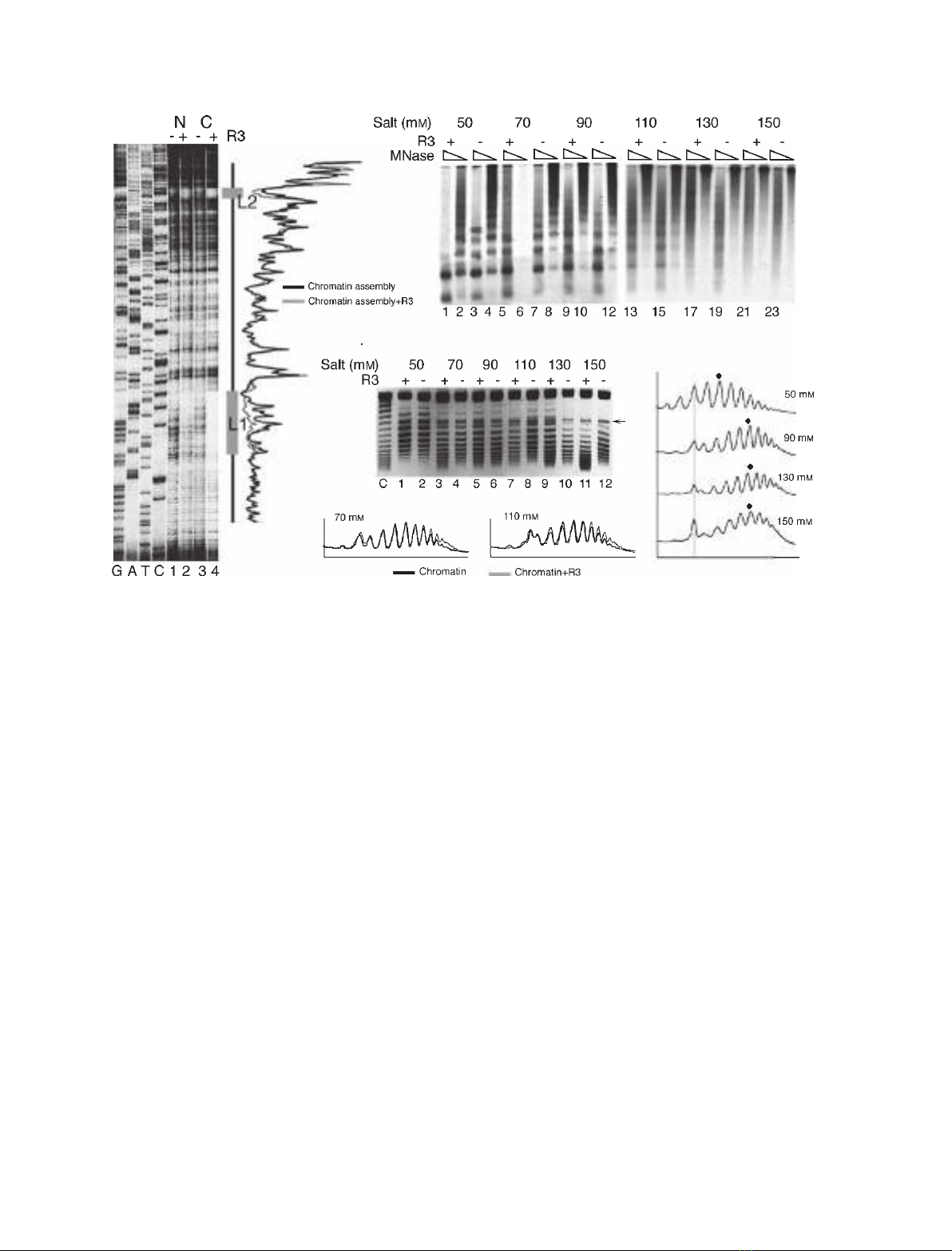
to the absence of R3 binding, but is probably related
to the reported effects of the ionic strength on the bulk
chromatin properties. As an increase in the ionic
strength is reported to increase the NRL [18], the total
number of uniformly spaced nucleosomes on a plasmid
DNA may decrease, which would result in a change in
the topological state of the plasmid. Therefore, we
confirmed the ionic strength effects on nucleosomal
density on plasmid DNA by the one-dimensional
supercoiling assay in the presence of chloroquin. The
plasmid in this assay was not relaxed prior to chroma-
tin assembly, as the S-190 extract is known to have
topoisomerase I activity, and assembly in this system
proceeds to completion. Therefore, under the condi-
tions of the gel run in Fig. 3C, all of the resolved
bands may be positively supercoiled topoisomers,
which differ by one linking number [30]. Comparison
of the profiles of the topoisomers (Fig. 3D) showed a
downward shift of the mean of the Gaussian distribu-
tion at different salt concentrations, denoting an
increase in the linking number of the DNA. As chloro-
quin introduces positive supercoils into the DNA, this
shift confirms that with increasing salt concentration,
there is a change in the superhelical density, which is
caused by a decrease in the number of nucleosomes
deposited over the plasmid DNA. In contrast to this,
the topoisomer profiles in the presence and absence of
R3 protein at different salt concentrations showed a
perfect overlap (Fig. 3E), suggesting that binding of
R3 at these ionic strengths did not change the nucleo-
some spacing further. Although the chromatin assem-
bly appeared to be better in the absence of R3
AB
C
E
D
Fig. 3. Influence of ionic strength and R3 binding on chromatin assembly. (A) Binding of R3 to its sites in the chromatin. High-resolution
DNaseI footprinting gel shows digestion profiles of the naked DNA (N) and chromatin assembly (C) in the presence and absence of the R3
protein. Chromatin was assembled with S-190 extract over plasmid pU6lac3 at a monovalent salt concentration of 150 mM. Comparison of
the profiles of lanes 3 and 4 in the right-hand panel shows protection due to R3 at L1 and L2 (gray boxes) but not between them. The pri-
mer was located 93 bp upstream of L1. GATC shows the sequencing ladders generated by the same primer. (B) MNase-resistant nucleo-
some ladders from chromatin assemblies were resolved on 1.25% agarose gels, Southern transferred, and probed with a primer that
hybridizes to the top strand, 53 bp downstream of L1. (C) One-dimensional supercoiling assay of chromatin assembly. Topoisomers were
resolved on a 1% agarose gel with 15 lMchloroquin present in the gel as well as the tank buffer. The arrow marks a band seen in every
lane, used as a reference. (C) Plasmid DNA control, which was not subjected to chromatin assembly. (D) Profiles of the topoisomer distribu-
tion of chromatin from lanes 2, 6, 10 and 12 in (C). The gray vertical line shows the alignment of peaks corresponding to the band marked
with an arrow in (C), and the dot marks the peak with the highest intensity in a profile. (E) Profiles of the topoisomer distribution in lanes 3
and 7 (chromatin with R3) are compared with those in lanes 4 and 8 (chromatin without R3) in (C).
R.-H. Pusarla et al. Mechanisms of nucleosome positioning
FEBS Journal 274 (2007) 2396–2410 ª2007 The Authors Journal compilation ª2007 FEBS 2399
(Fig. 3C), at 150 mmsalt, the significantly different
superhelical density (Fig. 3C) and presence of only
sparse nucleosomes in an MNase ladder assay (not
shown) suggest that the absence of positioned nucleo-
somes in this case is due to inefficient chromatin assem-
bly, rather than the loss of the boundary effect of R3.
Spacing between the positioned nucleosomes
does not change with ionic strength
The bulk chromatin is reported to show an increase in
NRL with increasing ionic strength [18]. Although this
may influence the spacing even between the positioned
nucleosomes, the IEL analysis in Fig. 2 suggested that
the relative location of the positioned nucleosomes
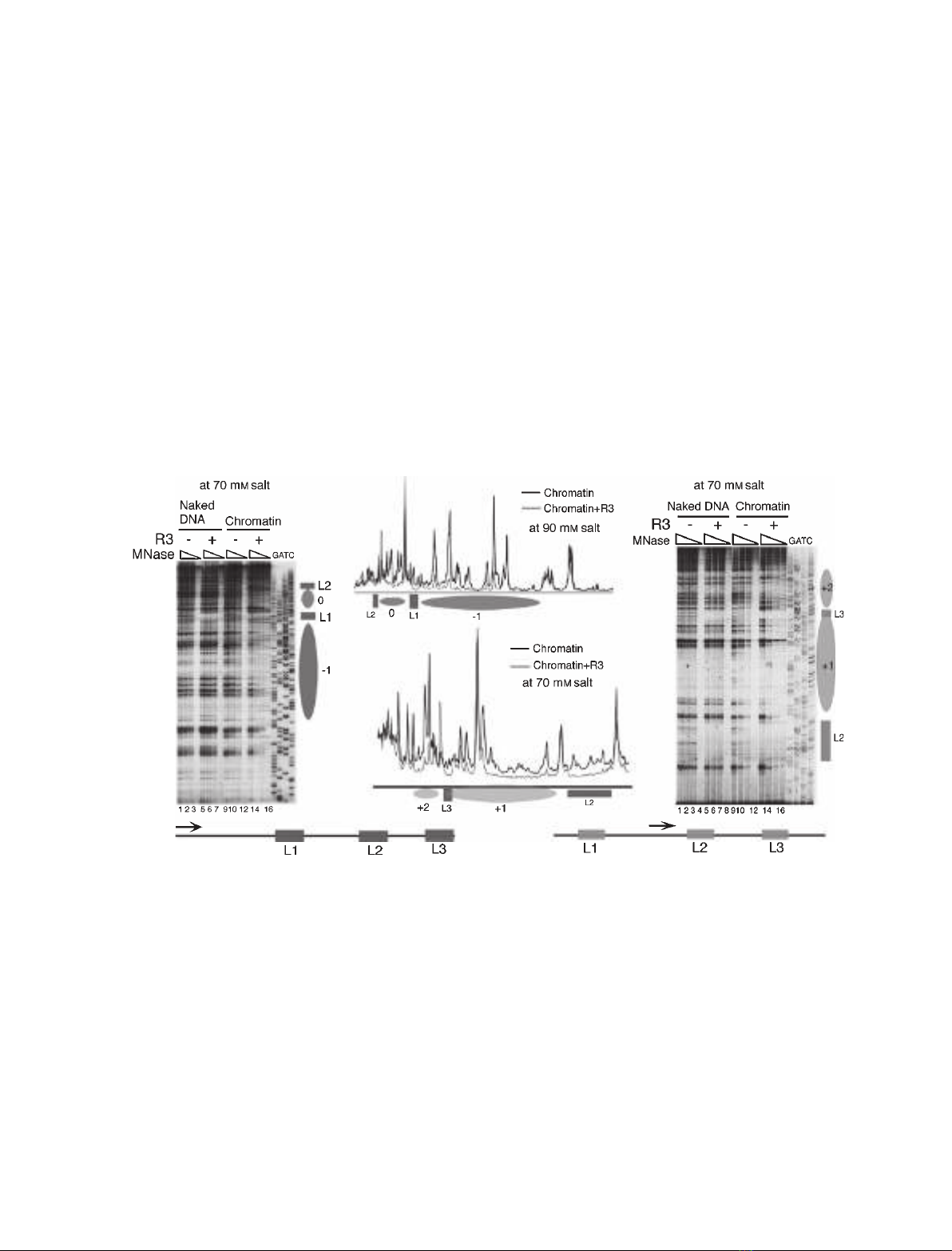
remains the same at every ionic strength. Numbering
the nucleosome between the operators 183 bp apart
as 0 (Fig. 2A), we used MNase footprinting to map
the positions of nucleosomes 0, )1 and + 1 by using
different primers to look at one nucleosome at a time
(Fig. 4). Translationally positioned nucleosomes
showed a clear 145 bp protection in the profile com-
parisons of the chromatin lanes with and without R3.
The + 1 nucleosome was found with its 5¢edge
located 10 bp downstream of the lac operator L2
(Fig. 4D), whereas the 3¢edge of nucleosome )1 was
found 10 bp upstream of the lac operator L1 (Fig. 4B)
at every ionic strength tested. Mapping of nucleosome
0 between the lac operators L1 and L2 showed its
location to be 25 bp downstream of L1 on the 5¢side,
and 12 bp upstream of L2 on the 3¢side. Similarly, the
location of the + 2 and )2 nucleosomes did not
change with changing ionic strength. This analysis
shows that the number of positioned nucleosomes
AB C
D
Fig. 4. Structural analysis of the pU6lac3 chromatin. High-resolution MNase footprinting was used to map the positioned nucleosomes over
plasmid pU6lac3 without or with bound R3. A seven-fold molar excess of R3 over operators was added at the start of the assembly. Aliqu-
ots of the same assembly were subjected to three levels of MNase digestion. Ellipses mark the nucleosomal size protections, and solid
boxes represent the R3 footprint and the lac operators. The positions of nucleosomes )1, 0, + 1 and + 2 are marked. (A) Mapping the posi-
tion of the nucleosomes in the presence of R3 by the primer extension footprinting of the chromatin assembled at 70 mMsalt. The primer
was located 196 bp upstream of the lac operator L1, hybridizing to the bottom strand, as depicted in the cartoon in the left-hand bottom cor-
ner. Lanes 1–8 show extension products of naked DNA digestions, and lanes 9–16 show chromatin samples. R3 was added to the samples
in lanes 5–8 and 13–16. GATC shows the sequencing ladder generated with the same primer. (B) Primer extension footprinting of the chro-
matin reconstituted at 90 mMsalt. A comparison of the profiles of the digested chromatin without and with R3 is shown. The primer was
the same as in (A). (C) Higher-resolution MNase footprinting of the chromatin with R3 bound or unbound shows that the lac operator L3 gets
included in the positioned nucleosome + 1. Lanes 1–8 show the naked DNA pattern with (lanes 5–8) and without (lanes 1–4) R3, and
lanes 9–16 show the digestion pattern of the chromatin assembled at 70 mMsalt (R3 was added to lanes 13–16). The primer was located
64 bp upstream of lac operator L2, hybridizing to the bottom strand as shown schematically in the right-hand bottom corner. GATC shows
the sequencing ladder generated by using the same primer. (D) A comparison of the profiles of lanes 9 and 13 in (C) is shown. Protection of
145 bp due to a positioned nucleosome + 1 overlaps with part of L3 occupied by R3.
Mechanisms of nucleosome positioning R.-H. Pusarla et al.
2400 FEBS Journal 274 (2007) 2396–2410 ª2007 The Authors Journal compilation ª2007 FEBS


























