
MINIREVIEW
Regulation of pyruvate dehydrogenase complex activity
in plant cells
Alejandro Tovar-Me
´ndez
1
, Jan A. Miernyk
1,2
and Douglas D. Randall
1
1
Department of Biochemistry, University of Missouri, Columbia, USA;
2
Plant Genetics Research Unit, USDA,
Agricultural Research Service, Columbia, USA
The pyruvate dehydrogenase complex (PDC) is subjected to
multiple interacting levels of control in plant cells. The first
level is subcellular compartmentation. Plant cells are unique
in having two distinct, spatially separated forms of the PDC;
mitochondrial (mtPDC) and plastidial (plPDC). The
mtPDC is the site of carbon entry into the tricarboxylic acid
cycle, while the plPDC provides acetyl-CoA and NADH for
de novo fatty acid biosynthesis. The second level of regula-
tion of PDC activity is the control of gene expression. The
genes encoding the subunits of the mt- and plPDCs are
expressed following developmental programs, and are
additionally subject to physiological and environmental
cues. Thirdly, both the mt- and plPDCs are sensitive to
product inhibition, and, potentially, to metabolite effectors.
Finally, the two different forms of the complex are regulated
by distinct organelle-specific mechanisms. Activity of the
mtPDC is regulated by reversible phosphorylation catalyzed
by intrinsic kinase and phosphatase components. An addi-
tional level of sensitivity is provided by metabolite control of
the kinase activity. The plPDC is not regulated by reversible
phosphorylation. Instead, activity is controlled to a large
extent by the physical environment that exists in the plastid
stroma.
Keywords: complex; chloroplast; enzymology; localization;
metabolic regulation; mitochondria; phosphorylation.
Introduction
The pyruvate dehydrogenase complex (PDC) is a multien-
zyme complex catalyzing the oxidative decarboxylation of
pyruvate to yield acetyl-CoA and NADH. The plant PDCs
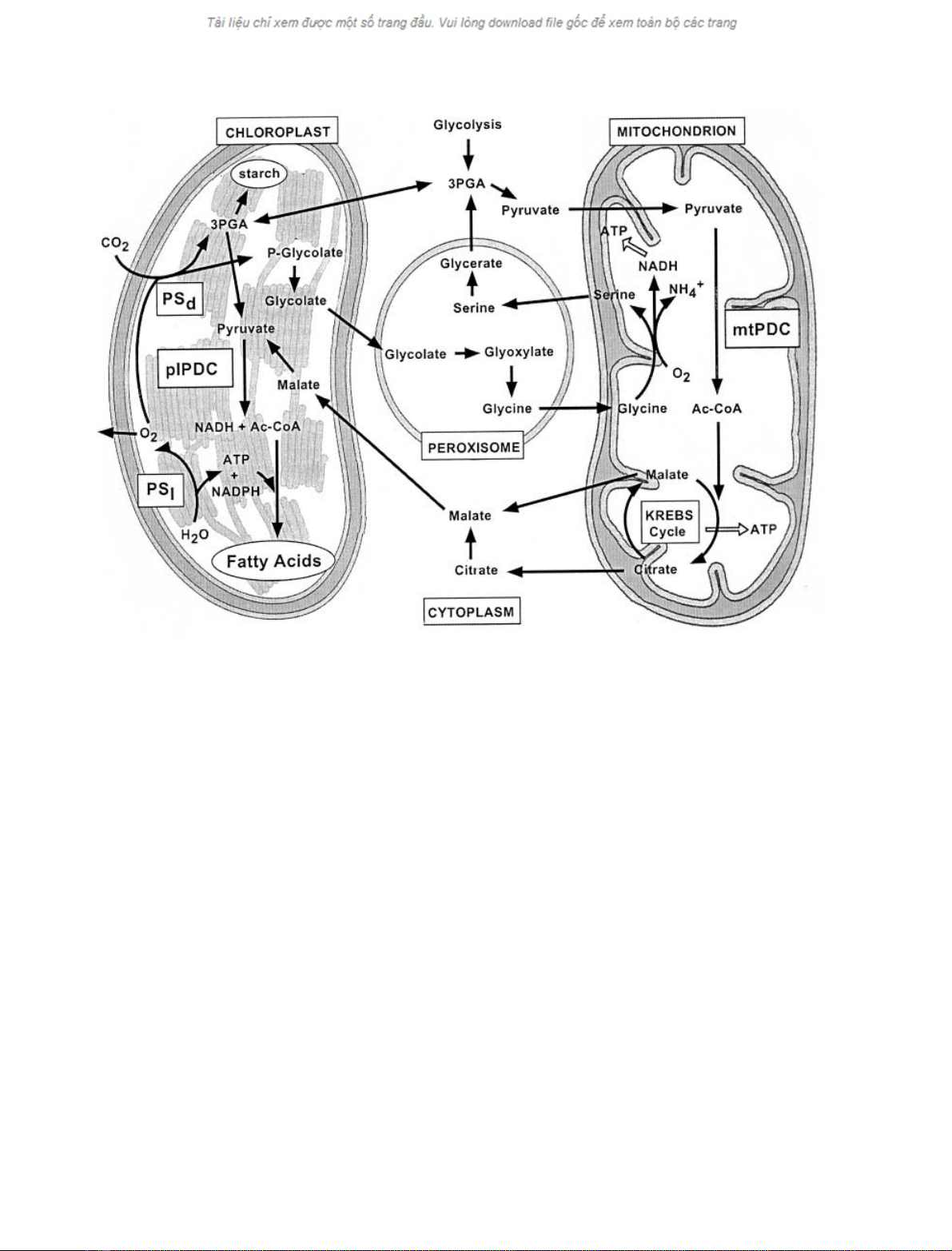
occupy strategic and overlapping positions in plant cata-
bolic and anabolic metabolism (Fig. 1). Similar to other
PDCs, the plant complexes contain three primary compo-
nents: pyruvate dehydrogenase (E1), dihydrolipoyl acetyl-
transferase (E2) and dihydrolipoyl dehydrogenase (E3). In
addition, mitochondrial PDC (mtPDC) has two associated
regulatory enzymes: pyruvate dehydrogenase kinase (PDK)
and phospho-pyruvate dehydrogenase phosphatase (PDP).
Here we briefly describe our current understanding of the
regulation of PDC activity in plant cells. Detailed descrip-
tions of the plant complexes are provided by more
comprehensive reviews [1–3].
Compartmentation of the PDC
It is widely believed that eukaryotic cells arose as the result
of phagotrophic capture of bacteria and subsequent sym-
biotic association. The progenitors of mitochondria are
thought to be a-proteobacteria [4], possibly related to
contemporary Rickettsia [5]. The plastids that are charac-
teristic of plant cells are thought to have been derived from a
single common primary symbiotic event with a cyanobac-
terium [6]. Subsequently, there was extensive gene migration
to the nucleus leaving both mitochondria and plastids as
semiautonomous organelles. Most mitochondrial and plas-
tidial proteins, including the subunits of the PDC, are
encoded within the nuclear genome of land plants, synthes-
ized in the cytoplasm and then post-translationally imported
into the organelles [3]. In nonplant eukaryotes the PDC is
exclusively localized within the mitochondrial matrix, and
serves as an entry point for carbon into the Krebs cycle. The
regulatory properties of mtPDC have been specialized to
minimize activity in an environment where ATP levels are
high. Plant cells contain an mtPDC that is closely related to
those of animal cells, but additionally contain a plastidial
form of the PDC (plPDC, Fig. 1) that is more closely
related to the PDC from cyanobacteria [3,7]. In contrast to
mtPDC, the regulatory properties of plPDC are specialized
to minimize the effects of an environment with high levels of
ATP. The physical environment within the chloroplast
stroma changes markedly during the light/dark transition,
and specialized regulatory mechanisms have evolved for
control of plPDC activity in the dark.
Mature plastids differentiate from proplastid progenitors
to serve specialized functions in different plant organs.
Plastid terminology is largely based upon pigmentation,
Correspondence to J. A. Miernyk, USDA/ARS, Plant Genetics
Research Unit, 108 Curtis Hall, University of Missouri,
Columbia, MO 65211, USA.
Fax: + 1 573 884 7850, Tel.: + 1 573 882 8167,
E-mail: miernykj@missouri.edu
Abbreviations: PDC, pyruvate dehydrogenase complex; mtPDC,
mitochondrial pyruvate dehydrogenase complex; plPDC,
plastidial pyruvate dehydrogenase complex; E1, pyruvate dehydro-
genase; E2, dihydrolipoyl acetyltransferase; and E3, dihydrolipoyl
dehydrogenase; PDP, phospho-pyruvate dehydrogenase phosphatase.
(Received 13 September 2002, accepted 29 November 2002)
Eur. J. Biochem. 270, 1043–1049 (2003) ÓFEBS 2003 doi:10.1046/j.1432-1033.2003.03469.x
with leucoplasts, etioplasts, chloroplasts and chromoplasts
being, respectively, unpigmented, pale yellow, green and
red/orange. The chlorophyll-containing green plastids
(chloroplasts) are the site of photosynthesis in autotrophic
plant cells. Plastids, regardless of pigmentation or degree of
differentiation, are the sole site of de novo fatty acid
biosynthesis in plant cells [8]. All forms of plastids contain
the plPDC, which provides the acetyl-CoA and NADH
necessary for fatty acid biosynthesis [9].
Recently it has been discovered that certain animal cell
parasites, such as Plasmodium spp., contain a type of
nonphotosynthetic plastid termed the apicoplast [10]. Pos-
sibly this type of plastid originated from an endosymbiotic
event involving a red algal cell. The fragmentary informa-
tion available indicates that red algal plPDCs are more
closely related to other plPDCs than to any mtPDC [3,7].
There is as yet no sequence information concerning red algal
mtPDC or plasmodial PDCs, but when this becomes
available it should provide us with additional phylogenetic,
evolutionary and regulatory insights.
Plastidial PDC
Based upon the results of cell-fractionation, it was proposed
that developing oilseeds contain a plastidial glycolytic
pathway in addition to the classical cytoplasmic glycolysis
[11]. It was additionally reported that these same plastids
contain a unique form of the PDC [12–14]. The plPDC from
developing castor endosperm has the same kinetic mechan-
ism as mtPDC, but has distinct catalytic and enzymatic
properties. It was later reported that green leaves from pea
seedlings also contain both mitochondrial and plastidial
forms of the PDC [15]. The occurrence of plPDC was briefly
controversial, however all of the subunits have now been
cloned [7,16,17] and their plastidial localization verified by
in vitro import studies [16,18] and confocal microscopy of
GFP-fusion proteins [19].
Similar to bacterial and mtPDC, the activity of plPDC is
sensitive to product inhibition by NADH and acetyl-CoA
[9,20]. Another property that is shared with bacterial PDCs
is that plPDC is not regulated by phosphorylation. Early
enzymatic studies of plPDC noted that the pH optimum
was significantly more alkaline than that of mtPDC, and
that higher Mg
2+
concentrations were necessary for maxi-
mal activity [9,12]. When plant leaves are shifted from dark
to light there is a rapid alkalinization of the chloroplast
stroma along with an increase in the free Mg
2+
concentra-
tion [21]. Both of these changes would activate plPDC.
De novo synthesis of fatty acids in green organs of plant cells
is light-driven and occurs exclusively within the plastids [8].
The plPDC provides acetyl-CoA and NADH for fatty acid
biosynthesis [9], so it is essential that PDC activity parallels
that of fatty acid biosynthesis. Thus, a unique mechanism
for regulating activity of plPDC activity has evolved based
Fig. 1. Compartmentalization of metabolism in plant cells. PS
l
, the light reactions of photosynthesis; PS
d
, the dark reactions of photosynthesis.
1044 A. Tovar-Me
´ndez et al.(Eur. J. Biochem. 270)ÓFEBS 2003
upon the physical conditions present in the chloroplast
stroma (Fig. 2). It is additionally possible that the activity of
plPDC [22] might be sensitive to light:dark changes in the
redox state of the chloroplast stroma [23] as are several
chloroplast regulatory enzymes [24].
Expression of plPDC
Expression of genes encoding the component enzymes of
plPDC is responsive to developmental and physiological
cues. The level of plE1bmRNA expressed in A. thaliana
siliques increased to a peak six to seven days after
flowering, then decreased with seed maturity [25]. This
pattern of developmental expression is parallel to that of
plastidial acetyl-CoA carboxylase, consistent with a role for
both enzymes in seed oil synthesis and accumulation [25].
The importance of plPDC in seed oil synthesis has been
further supported by results from both digital Northern
[26] and microarray [27] analyses of developing A. thaliana
seeds.
In addition to developing seeds, it has been reported that
there were high levels of expression of plE1b[25], plE2 [16],
and plE3 [17] in A. thaliana flowers. However, when the
b-glucuronidase (GUS) reporter gene was fused to the
A. thaliana plE3 promoter, and this chimera expressed in
tobacco plants, high levels of expression were seen in
developing seeds and mature pollen grains while low levels
were present in young leaves and flowers [19]. This result
suggests the previously reported elevated levels of plPDC
subunit expression in flowers might instead reflect mRNAs
present in the pollen.
Mitochondrial PDC
Product inhibition
As with their mammalian and microbial counterparts, plant
PDCs employ a multisite ping-pong kinetic mechanism. The
forward reaction is irreversible under physiological condi-
tions, but activity is sensitive to product inhibition by
NADH and acetyl-CoA. The K
i
values for NADH (20 l
M
)
and acetyl-CoA (20 l
M
) are within the physiological
concentration range [28]. While the results from in vitro
studies suggest that NAD
+
/NADH is the more important
regulator, results from analyses using isolated intact mito-
chondria suggest that acetyl-CoA/CoA can also have a
significant regulatory influence because of the small size of
the total CoA pool [29].
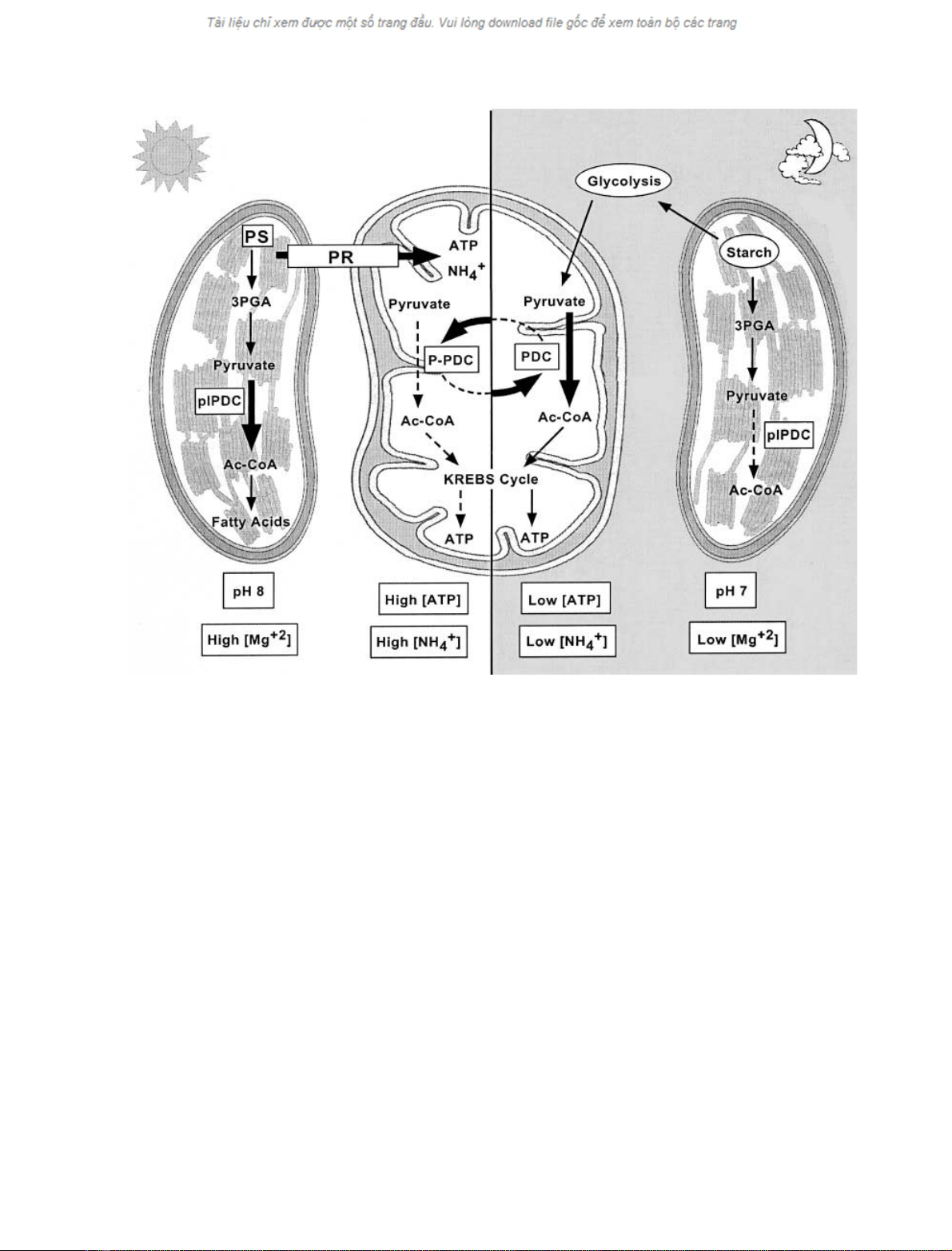
Fig. 2. Schematic overview of the regulation of pyruvate dehydrogenase complex activity in autotrophic plant cells. Distinct regulatory mechanisms
control the activity of mtPDC in the light and plPDC in the dark. PS, photosynthesis; PR, the photorespiratory pathway; PDC, the pyruvate
dehydrogenase complex; P-PDC, the phosphorylated (inactive) form of PDC.
ÓFEBS 2003 Regulation of PDC activity in plant cells (Eur. J. Biochem. 270) 1045


























