
RESEARCH ARTICLE Open Access
High-throughput SNP genotyping in the highly
heterozygous genome of Eucalyptus: assay
success, polymorphism and transferability across
species
Dario Grattapaglia
1,2*
, Orzenil B Silva-Junior
1
, Matias Kirst
3
, Bruno Marco de Lima
1,4
, Danielle A Faria
1
and
Georgios J Pappas Jr
1,2
Abstract
Background: High-throughput SNP genotyping has become an essential requirement for molecular breeding and
population genomics studies in plant species. Large scale SNP developments have been reported for several
mainstream crops. A growing interest now exists to expand the speed and resolution of genetic analysis to
outbred species with highly heterozygous genomes. When nucleotide diversity is high, a refined diagnosis of the
target SNP sequence context is needed to convert queried SNPs into high-quality genotypes using the Golden
Gate Genotyping Technology (GGGT). This issue becomes exacerbated when attempting to transfer SNPs across
species, a scarcely explored topic in plants, and likely to become significant for population genomics and inter
specific breeding applications in less domesticated and less funded plant genera.
Results: We have successfully developed the first set of 768 SNPs assayed by the GGGT for the highly
heterozygous genome of Eucalyptus from a mixed Sanger/454 database with 1,164,695 ESTs and the preliminary
4.5X draft genome sequence for E. grandis. A systematic assessment of in silico SNP filtering requirements showed
that stringent constraints on the SNP surrounding sequences have a significant impact on SNP genotyping
performance and polymorphism. SNP assay success was high for the 288 SNPs selected with more rigorous in silico
constraints; 93% of them provided high quality genotype calls and 71% of them were polymorphic in a diverse
panel of 96 individuals of five different species.
SNP reliability was high across nine Eucalyptus species belonging to three sections within subgenus Symphomyrtus
and still satisfactory across species of two additional subgenera, although polymorphism declined as phylogenetic
distance increased.
Conclusions: This study indicates that the GGGT performs well both within and across species of Eucalyptus
notwithstanding its nucleotide diversity ≥2%. The development of a much larger array of informative SNPs across
multiple Eucalyptus species is feasible, although strongly dependent on having a representative and sufficiently
deep collection of sequences from many individuals of each target species. A higher density SNP platform will be
instrumental to undertake genome-wide phylogenetic and population genomics studies and to implement
molecular breeding by Genomic Selection in Eucalyptus.
* Correspondence: dario@cenargen.embrapa.br
1
EMBRAPA Genetic Resources and Biotechnology - Estação Parque Biológico,
final W5 norte, Brasilia, Brazil
Full list of author information is available at the end of the article
Grattapaglia et al.BMC Plant Biology 2011, 11:65
http://www.biomedcentral.com/1471-2229/11/65
© 2011 Grattapaglia et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative
Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and
reproduction in any medium, provided the original work is properly cited.

Background
High-throughput, high density SNP genotyping has
become an essential tool for QTL mapping, association
genetics, gene discovery, germplasm characterization,
molecular breeding and population genomics studies in
several crops and model plants [1-7]. The abundance of
Single Nucleotide Polymorphisms (SNPs) in plant gen-
omes together with the rapidly falling costs and
increased accessibility of genotyping technologies, have
prompted an increasing interest to develop panels of
SNP markers to expand resolution and throughput of
genetic analysis in less-domesticated plant species with
uncharacterized genomes such as those of orphan crops
[8], forest [9-12] and fruit trees [13-15].
Two main strategies have been employed to identify
SNPs in plants: utilization of EST sequence information
to direct targeted amplicon resequencing and, more
recently, next generation sequencing (NGS) technologies
coupled or not to genome complexity reduction meth-
ods [16]. Amplicon resequencing of stretches of target
genes is carried out in a germplasm panel that is rele-
vant to the downstream applications and sufficiently
large to avoid ascertainment bias. SNPs are mined in
the resulting sequences and then assays are designed
focusing on those particular SNPs. This strategy,
although labor intensive, has been successful when the
goal is to develop a moderate number of assayable SNPs
[16]. High throughput NGS and direct in silico SNP
identification now provide a very effective alternative to
amplicon resequencing for SNP development in plants
[17]. Thousands of SNPs can be readily identified given
that sequences are obtained from an adequately large
representation of individuals with sufficiently redundant
genome coverage. Complexity reduction strategies such
as using cDNA libraries [18,19], AFLP derived represen-
tations [20], reduced representation libraries generated
by restriction enzyme digestion and fragment selection
[2,21], microarray-based [22] or in-solution [23]
sequence capture, and additional target enrichment stra-
tegies [24] can be used to obtain the necessary sequence
depth when the objective is to develop SNP based mar-
kers in specific genes or regions of the genome. Multi-
plexed bar-coded sequencing of such reduced genomic
representations optimizes costs of SNP identification by
increasing coverage and genotypic representation in the
target regions [24-26]. Clearly the prospects are that
sequence abundance and quality for SNP identification
will no longer be a limiting factor for any plant genome.
A number of SNP genotyping technologies were
developed in recent years mostly geared toward assaying
human SNP variation. Among those that have been
used in plant genetics, the Golden Gate Genotyping
Technology (GGGT) developed by Illumina has consis-
tently been reported as a reliable technology, displaying
high levels of SNP conversion rate and reproducibility
[16]. This assessment, initially reported for large scale
human genotyping, has been corroborated in plant spe-
cies including autogamous crops with low nucleotide
diversity (0.2% to 0.5%) [3,27-29] and outbred species
with much higher sequence diversity typically ≥2%
[9-13]. In highly heterozygous genomes, the develop-
ment of GGGT SNP assays has been carried out mainly
by amplicon resequencing targeting specific genes. This
approach has been practical in conifers using haploid
megagametophyte tissue [30,31] and poplar for which a
reference genome is available [12]. If attempted for large
scale SNP development, however, this approach would
be technically challenging for most outbred plant gen-
omes due to the high levels of nucleotide diversity and
additional indel variation as shown in earlier attempt for
grape [32]. Direct SNP development from large in silico
sequence resources will likely be the best approach for
the highly heterozygous genomes of the majority of
undomesticated plant species.
Irrespective of the method used to develop SNP mar-
kers in heterozygous genomes - direct in silico or tar-
geted amplicon re-sequencing - challenges are faced in
later steps when attempting to convert queried SNPs
into high-quality genotypes. Particularly for the develop-
ment of GGGT assays based on hybridization of allele
and locus specific oligonucleotides, constraints have to
be placed on the sequences flanking the target SNP
[33]. A robust diagnosis of sequence variation in the
vicinity of the target SNPs will depend largely on
sequence coverage, sequence quality [34] and origin of
sequences as far as the number and relatedness of indi-
viduals surveyed for SNP discovery. These issues will
become increasingly exacerbated when attempting to
transfer SNP assays across species within the same
genus. Still a rarely explored topic in plants [13,30,35],
the assessment of inter-specific transferability of SNPs
will likely be an important subject for population geno-
mics and inter specific breeding applications in less
domesticated and less funded plant genera.
Species of Eucalyptus are currently planted in more
than 90 countries and are well known for their fast
growth, straight form, valuable wood properties and
wide adaptability [36]. Eucalyptus subgenus Symphyo-
myrtus, includes the majority of the twenty or so com-
mercially planted species. E. globulus has been the top
choice for plantations in temperate regions. Tropical
Eucalyptus forestry, on the other hand, is based on
interspecific hybrid breeding and clonal propagation
with E. grandis as the pivotal species [36]. Molecular
marker technologies have allowed a significant progress
in the genetics and breeding of this vast genus that
includes over 700 species [36]. Genetic analyses with
molecular markers were key to settle phylogenetic issues
Grattapaglia et al.BMC Plant Biology 2011, 11:65
http://www.biomedcentral.com/1471-2229/11/65
Page 2 of 18

[37], manage breeding populations [38] build linkage
maps [39-41] and identify QTLs for important traits
[42-45]. Nonetheless, more extensive genome coverage,
higher throughput and improved inter specific transfer-
ability of current genotyping methods are necessary to
increase resolution and speed for a variety of applica-
tions. A DArT array delivering around 3,000 to 5,000
dominant markers for mapping and population analyses
was recently reported [46]. SNP developments in species
of the genus have targeted specific candidate genes gen-
erating a few tens SNPs for specific association genetics
studies [47,48]. However, large scale SNP arrays devel-
opments for Eucalyptus are yet to come. Due to their
recent domestication, large population sizes and outbred
mating system, species of Eucalyptus are among the
ones with the highest frequency of SNPs reported in
woody plant species and possibly in plants in general,
with up to 1 SNP every 16 bp [49]. While a bonus for
overall SNPs identification, such high nucleotide diver-
sity, both within and among species, could represent an
obstacle for the development of large sets of robust and
polymorphic sets of Golden Gate assayable SNPs across
species.
We are interested in developing genome-wide paralle-
lized genotyping methods to be used for the operational
implementation of Genomic Selection in Eucalyptus
hybrid breeding, population genomics and phylogenetic
studies in natural populations of the genus. The upcom-
ing availability of a reference genome for Eucalyptus
grandis and the rapid evolution of high throughput
sequencing technologies will foster the buildup of large
sequence dataset from many individuals, a valuable
resource for the development of large collections of
SNPs for the genus. In anticipation to this time, we
used a 1.2 million mixed EST dataset including Sanger
and 454 sequences from multiple Eucalyptus species
and individuals to: (1) develop and validate an initial
collection of genome-wide SNPs for Eucalyptus derived
exclusively from in silico EST sequence data from unre-
lated individuals of different species; (2) assess the effect
of increasingly stringent in silico SNP identification and
design parameters on the reliability and polymorphism
of SNP genotyping in species of Eucalyptus using the
Golden Gate Genotyping Technology (GGGT); (3) eval-
uate SNPs transferability across eleven species of Euca-
lyptus and polymorphism in the five main planted
species worldwide. Information on all SNPs discovered
and validated in the present study is provided.
Results
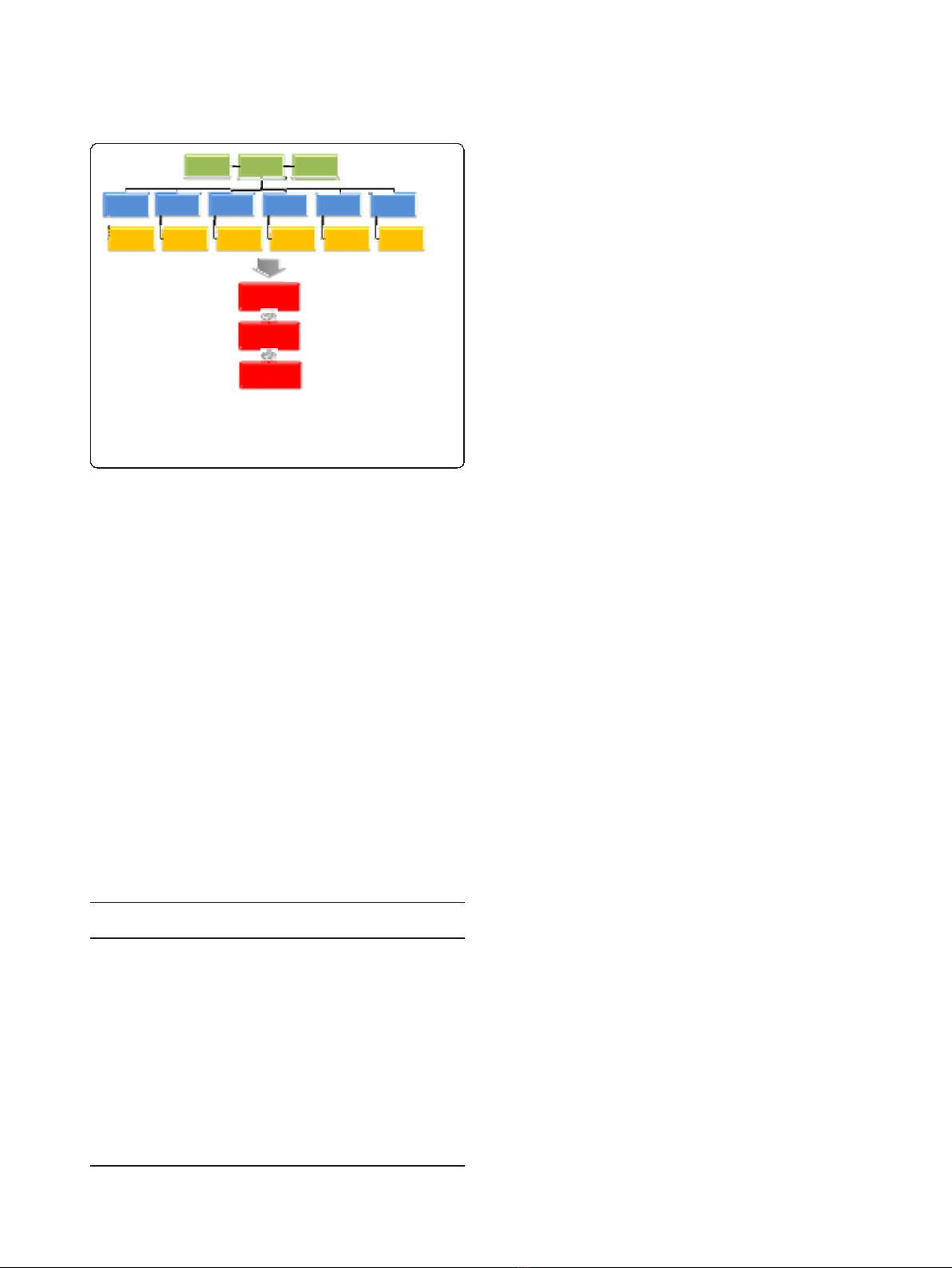
EST clustering, contig assembly and SNP discovery
pipeline
ESTs for six different species of Eucalyptus were used in
this study to maximize the sampling of DNA sequence
variation across species, although only a portion was
retained for assembly after applying several quality fil-
ters. From a total of 136,041 Sanger-derived ESTs,
78,087 of them (57.4%) were further processed. Similar
percentage was retained out of the 1,028,654 454-
derived ESTs (60.7%) (Table 1). The majority of the
Sanger reads and all 454 reads were obtained from
E. grandis, the pivotal species in most tropical breeding
programs, totaling 94% of the available ESTs before
assembly and 96% after assembly, i.e. effectively used for
SNP discovery. A two-step EST-assembly strategy was
used: clustering performed at the species and sequen-
cing technology levels followed by using the MIRA 2
assembler (Whole Genome Shotgun and EST Sequence
Assembler) to consolidate the contigs and singletons
from the previous step into a final EST assembly. After
the MIRA assembly 48,973 contigs were obtained. Only
those contigs formed by five or more ESTs were consid-
ered in this analysis to mitigate the limitations of align-
ment depth in SNP detection, thus resulting in 17,703
usable contigs (36.15% of the total). From this contig
set, SNPs were predicted using the program PolyBayes.
Only SNPs with high probability (P
SNP
≥0.99) were
selected, totaling 162,141 potentially polymorphic sites
(Figure 1).
In silico selection of genome-wide SNP
Five sequential filters were applied to the 162,141 candi-
date genome-wide SNPs for GGGT assay design from
F0 (less stringent) to F4 (most stringent) (see Methods).
When the filtering stringency increased from F0 to F4,
the number of SNPs surviving selection in silico
decreased abruptly. A total of 66,254 SNPs (40.6%) were
selected that had ≥5 reads on the SNP position and a
minimum of one read with the alternative base. This
number dropped to 21,944 (13.5%) when an in silico
MAF ≥0.2 constraint was applied and to 10,032 (6.2%)
whenatleastoneESTfromthemoredistantspeciesE.
globulus or E. gunnii was required in the contig. When
the filter requiring flanking sequence conservation was
applied, the number of SNPs selected dropped even
Table 1 Summary of the EST assembly for SNP discovery
Sequencing
technology
Eucalyptus
species
# sequences used
for clustering
# sequences in
the assembly
Sanger E. grandis 67,635 50,720
E. globulus 30,260 10,088
E. urophylla 7,755 4,387
E. gunnii 19,586 7,018
E. pellita 9,679 4,959
E. tereticornis 1,126 1,095
454 E. grandis 1,028,654 623,922
TOTAL 1,164,695 702,009
Grattapaglia et al.BMC Plant Biology 2011, 11:65
http://www.biomedcentral.com/1471-2229/11/65
Page 3 of 18
further to a final number of only 1,329 when a cutoff of
60 bases with no additional SNP on each side of the tar-
get SNP was stipulated. The number of unigene contigs
retained along the filters also dropped significantly from
an initial number of 17,703 to a mere 998 when all fil-
tering constraints were applied (Table 2). Overall the
proportion of SNPs with ADT (Assay design Tool)
score greater than 0.6, i.e. SNPs with a high likelihood
to be converted into a successful genotyping assay, was
around 95%, irrespective of the filtering treatments. For
example, by applying only filter F0, 598 SNPs out of 621
had ADT score ≥0.6; similarly, with filter F4, 525 out of
547 SNPs had ADT score ≥0.6. The proportion of SNPs
with ADT score ≥0.9 was between 50 and 53% again
showing no impact of the filtering treatments (Table 2).
For bench validation only SNPs with ADT score ≥0.8
were selected. A list of the 696 genome-wide SNPs
selected and tested by the Golden Gate assay is available
in Additional file 1.
SNP discovery in pre-determined candidate genes
From a list of 42 candidate genes selected from the lit-
erature as being putatively associated with relevant
wood phenotypes in Eucalyptus (see Material and Meth-
ods), only in 20 of them SNPs were found that matched
the minimum requirements of having ≥2readswith
alternative bases at the SNP position and at least 60
bases of flanking sequence on each SNP side. For these
20 genes, a total of 175 SNPs were discovered and 72
were included in the bead array for downstream valida-
tion. These 72 SNPs were selected to assay at least one
SNP in each one of the 20 genes and in those genes
where several SNPs were available, SNPs that were
derived from a contig with at least one read coming
from E. globulus or E. gunnii and distantly positioned
along the contig were selected. These 72 SNPs assayed
in candidate genes are available as a separate spread-
sheet in Additional file 1.
SNP genotyping reliability
The distributions of the proportions of SNPs in increas-
inglymorereliableclassesasmeasuredbytheGene-
Call50 and GeneTrain scores for each in silico filter
level were plotted (Figure 2). The relative distribution of
the broken bars histograms corresponding to increasing
levels of reliability suggests that when progressively
more stringent in silico SNP selection requirements are
applied from F0 to F4, larger proportions of SNPs with
higher GeneTrain and GC50 scores were obtained. For
SNPs in pre-determined candidate genes (CG) the pro-
portions of SNPs at the lower ends of the distribution
of GC50 and GeneTrain scores were larger reflecting
the less stringent in silico selection applied in these
cases (Figure 2). SNPs developed in specific candidate
genes for which limitations existed regarding the num-
ber of available EST reads, generally showed a slightly
lower performance in all measured parameters of relia-
bility even when compared to SNPs developed only
applying filter F0. The proportion of SNPs with call rate
rate ≥95% was only 80.6%, the average GeneTrain
scorewasthelowestat0.61,andtheproportionof
SNPs with GeneTrain and GC50 scores ≥0.40 was less
than 90%. However no difference was seen in the pro-
portion of polymorphic SNPs in relation to the more
stringent in silico filtering levels. Because SNPs in can-
didate genes were mined without observance of any
specific in silico filtering level besides the most funda-
mental one (see methods), they were not included in
the subsequent comparative analyses of the in silico fil-
tering parameters.
Genolyptus
101,240 ESTs
NCBI Genbank
34,801 ESTs
E. grandis
1,096,289 ESTs
32,473 contigs
642,169 singlets
E. globulus
30,260 ESTs
3,578 contigs
6,330 singlets
E. gunnii
19,586 ESTs
3,020 contigs
3,998 singlets
E. pellita
9,679 ESTs
1,775 contigs
3,184 singlets
E. urophylla
7,755 ESTs
1,194 contigs
3,193 singlets
E. tereticornis
1,126 ESTs
30 contigs
1,065 singlets
NCBI SRA
1,028,654 ESTs
48,973 contigs
17,703 contigs
162,141
Polybayes SNPs
ESTs grouped by species
Clustering and assembl
y
EST assembly with MIRA
Selection of contigs with ш5 reads
SNP detecion with Polybayes
ES
Figure 1 Flowchart with the output results of the EST
clustering, contig assembly and SNP discovery pipeline prior
to applying SNP filtering and selection for the GGGT assay
design.
Table 2 Summary of the in silico SNP development
procedure using increasingly stringent SNP selection and
design requirements (F0 through F4) (see methods for
details)
In silico SNP performance
assessment
F0 F1 F2 F3 F4
# of SNPs 66,254 21,944 10,032 3,187 1,329
# of contigs with SNPs 9,579 5,058 2,057 1,651 998
# of SNPs submitted to the
ADT
621 605 583 367 547
# of SNPs with ADT Score ≥
0.6
598 572 557 353 525
% of SNPs with ADT Score ≥
0.6
96.3 94.5 95.5 96.2 96.0
# of SNPs with ADT Score ≥
0.9
314 316 297 177 291
% of SNPs with ADT Score ≥
0.9
50.6 52.2 50.9 48.2 53.2
# of SNPs tested by the GGGT 96 96 108 108 288
Grattapaglia et al.BMC Plant Biology 2011, 11:65
http://www.biomedcentral.com/1471-2229/11/65
Page 4 of 18

F4
ALL 0Ͳ 0.2
0.2Ͳ 0.4
0.4Ͳ 0.6
F0
F1
F2
F3
F4
0.6Ͳ 0.8
0.8Ͳ 1.0
n
eTrainScore
0% 20% 40% 60% 80% 100%
CG
Ge
n
F4
ALL 0Ͳ 0.2
0.2Ͳ 0.4
0.4Ͳ 0.6
0.6Ͳ 0.8
(a)
CG
F0
F1
F2
F3
0.8Ͳ 1.0
e
neCall50Score
0% 20% 40% 60% 80% 100%
CG
G
e
F4
ALL 0Ͳ 0.05
0.05Ͳ 0.10
0.10Ͳ 0.15
0.15Ͳ 0.20
020
Ͳ
025
(b)
CG
F0
F1
F2
F3
0
.
20
0
.
25
0.25Ͳ 0.30
0.30Ͳ 0.35
0.35Ͳ 0.40
0.40Ͳ 0.45
0.45Ͳ 0.50
MAF
0% 20% 40% 60% 80% 100%
(c)
Figure 2 Distribution of the percentages of SNPs across classes of (a) GeneTrain Score; (b) GeneCall50 Score and (c) Minimum Allele
Frequency (MAF) . Broken bars histograms are presented for all 768 SNPs together (ALL) and for each SNP category within the 696 genome-
wide SNPs selected by the different in silico filtering levels (F0 through F4 - see methods) and the 72 candidate gene (CG) SNPs.
Grattapaglia et al.BMC Plant Biology 2011, 11:65
http://www.biomedcentral.com/1471-2229/11/65
Page 5 of 18


























