
J. Sci. Dev. 2009, 7 (Eng.Iss.1): 9 - 16 HA NOI UNIVERSITY OF AGRICULTURE
9
Application of PCR to identify the transgene
(Arabidopsis thaliana allene oxide synthase 2) in
transgenic rice plants
Ứng dụng PCR để phát hiện sự có mặt của gen AtAOS 2
trong genome của lúa đã được chuyển gen
Nguyen Thi Thuy Hanh 1, Han Oksoo 2
1Faculty of Agronomy, Hanoi University for Agriculture
2College of Agriculture and Life sciences, Chonnam National University, South Korea
TÓM TẮT
Allene oxide synthase (AOS) là một enzyme quan trọng trong số các enzyme tham gia vào quá
trình tổng hợp axit jasmonic thông qua con đường sinh hoá octadecanoid. Ở thực vật, axit jasmonic
đóng vai trò then chốt trong việc hình thành các phản ứng đáp lại các tác động của môi trường trong
và ngoài tế bào. Để làm tăng hàm lượng enzyme allene oxide synthase nội sinh ở lúa, gen allene
oxide synthase 2 của Arabidopsis thaliana (AtAOS 2) đã được chuyển vào genome của lúa (Oryza
sativa L.Japonica cv. Nakdong). Phản ứng PCR đã được sử dụng để phát hiện sự có mặt của gen
AtAOS2 trong genome của lúa đã được chuyển gen. Cụ thể là phản ứng PCR đã được sử dụng để
phát hiện gen tổng hợp chất kháng sinh hygromycin, promoter CaMV35S (promoter này nằm ngay
phía trước gen AtAOS2 trong cấu trúc của vector pCAMBIA1201/AtAOS) và gen AtAOS 2 trong
genome của lúa.
Từ khoá: Allene oxide synthase, axit jasmonic, con đường sinh hoá otadecanoid, thực vật
chuyển gen.
SUMMARY
Allene oxide synthase (AOS) is an important regulator that steers the octadecanoid pathway
toward jasmonic acid (JA) synthesis. Plant jasmonic acid (JA) plays pivotal roles regulating cellular
responses against environmental cues, including the innate immune response(s). The Arabidopsis
thaliana allene oxide synthase 2 (AtAOS 2) gene was introduced into rice (Oryza sativa L.Japonica
cv.Nakdong) genome. The polymeraza chain reactions (PCRs) were applied to indentify the transgene
- AtAOS 2 in transgenic rice plants. A set of PCRs was carried out to detect the hygromycin resistance
gene, CaMV35S promoter (the pomoter located infront of the AtAOS 2 gene in pCAMBIA1201/AtAOS
binary vector) and the AtAOS 2 gene in the transgenic rice genome.
Key words: Allene oxide synthase, jasmonic acid, octadecanoid pathway, transgenic plants.
1. INTRODUCTION
Plants have evolved defense strategies to
protect themselves from various abiotic and biotic
stresses. One of the most prominent defense
mechanisms is the octadecanoid pathway, which
leads to jasmonic acid (JA) biosynthesis. JA is a
genuine plant hormone that has various
physiological functions, including growth control,
senescence promotion, responses to wounding and
drought, and defenses against insects and pathogens
(Schaller et al., 2005). Furthermore, JA signaling
interacts with signaling of other plant hormones such
as ethylene and salicylic acid to form a complex
signaling network (Devoto et al., 2005). JA is
synthesized through the octadecanoid pathway
(Figure 1). In the octadecanoid pathway, 13 (S) -
HPOT is the pivotal intermediate, from which
several metabolic pathways branch out. For the JA
synthesis, 13 (S) - HPOT is converted to an unstable
allene oxide, 12,13 - epoxy - 9Z, 11E, 15Z -
octadecatrienoic acid (12,13 - EOT), by an allene
oxide synthase (AOS). The allene oxide is readily
converted to cis-(+)-OPDA by allene oxide cyclase
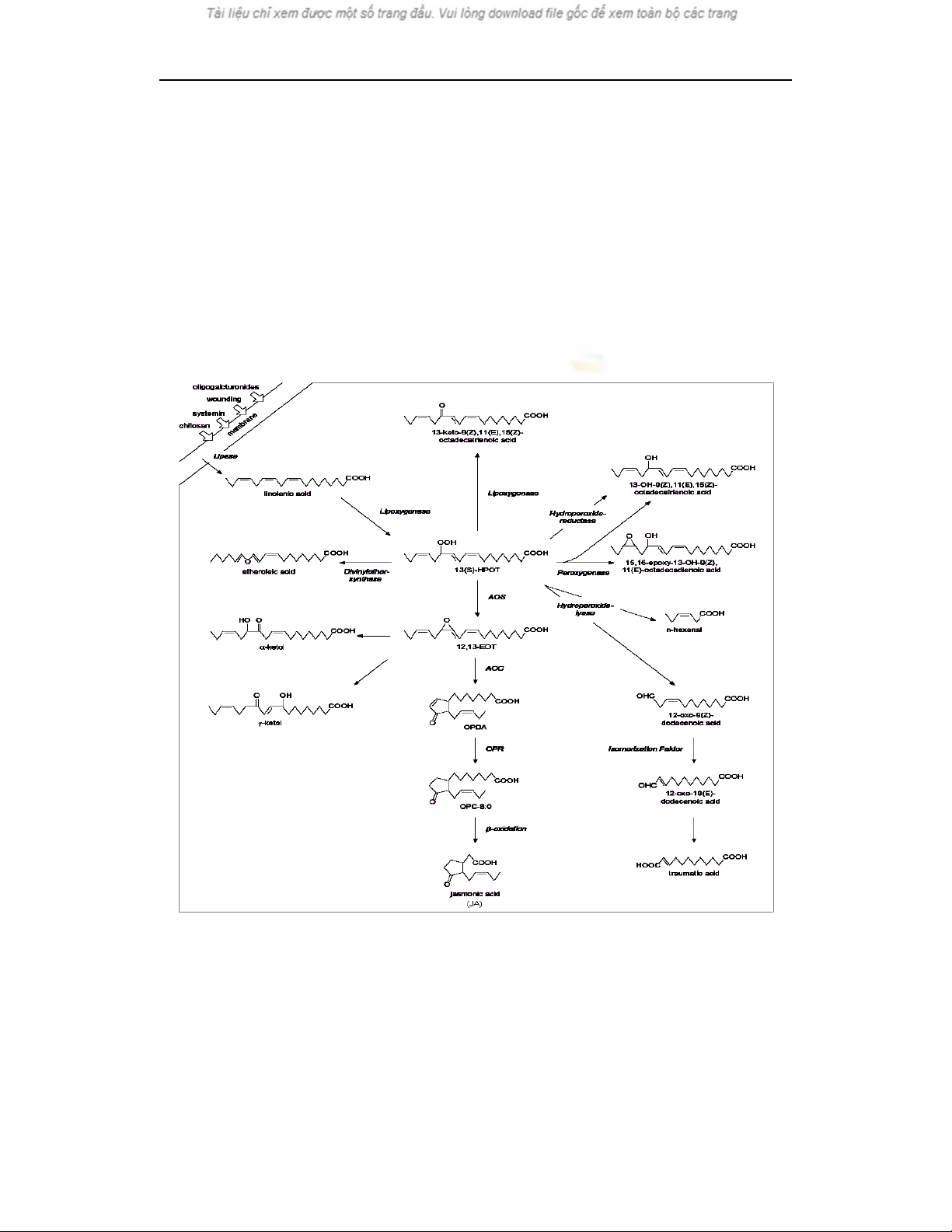
Application of PCR to identify the transgene (Arabidopsis thaliana allene oxide synthase2)...
10
(AOC); cis-(+)-OPDA is further metabolized
through reduction and β-oxidation to JA. On the
other hand, 13(S)-HPOT can also be metabolized by
other enzymes, including hydroperoxide lyase,
peroxygenase, hydroperoxide reductase, and divinyl
ether synthase. A typical function of these branch
pathways is defence against insect and pathogens
(BlÐe, 2002). Therefore, AOS is an important
regulator that steers the octadecanoid pathway to JA
synthesis, thus affecting the synthesis of all JA-
related compounds (Kongrit et al., 2007).
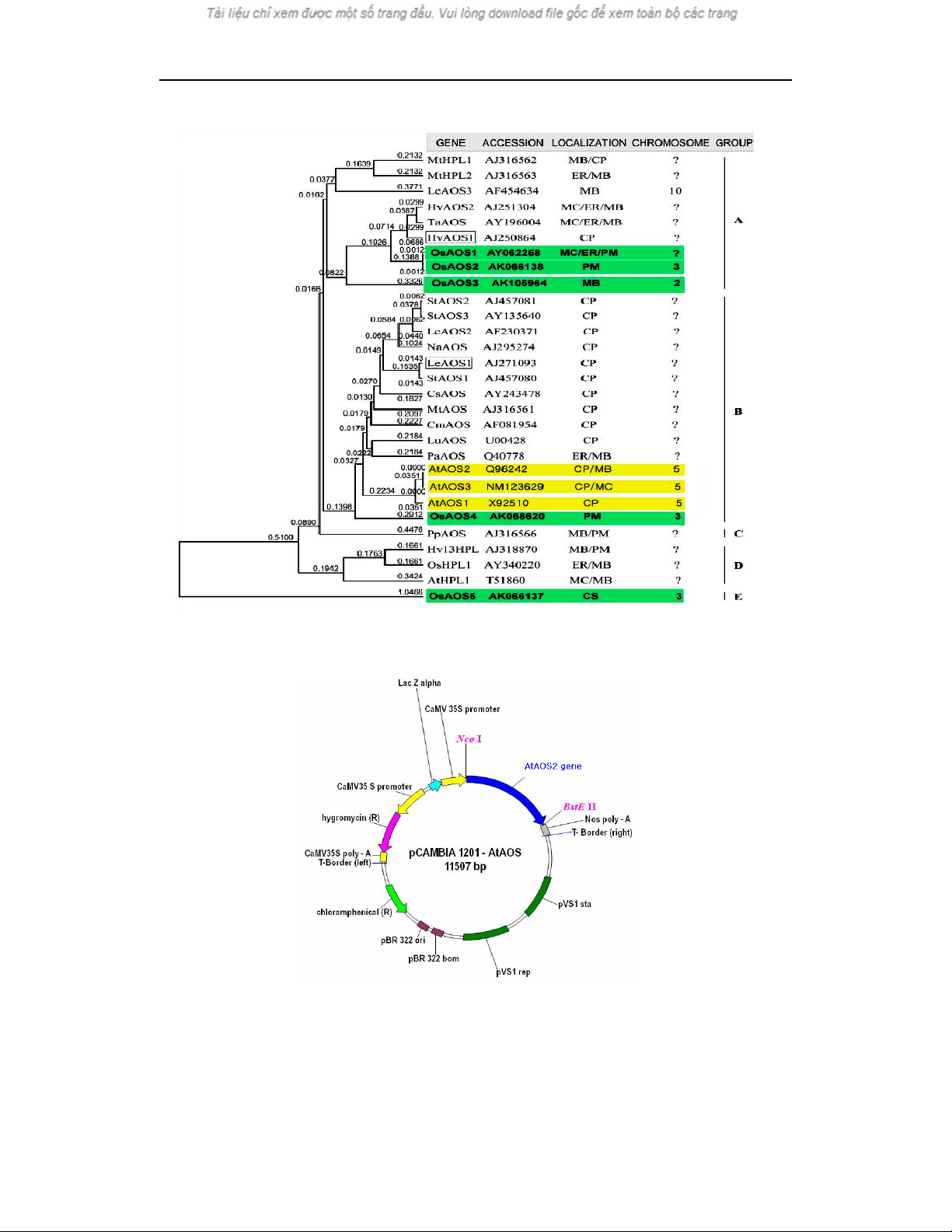
Rice AOS (OsAOS) and Arabidopsis
thaliana AOS belong to the CYP74A subfamily.
A database search indicates that, besides the
characterized OsAOS1 (Agrawal et al., 2004),
there are at least four additional OsAOS
(OsAOS2-5) genes in the rice genome (Figure 2).
Most of the rice AOS genes are located on
chromosome 3, except for the OsAOS3 gene, that
is located on chromosome 2. The OsAOS proteins
show putative localization sites such as
mitochondria (MC), endoplasmic reticulum (ER),
plasma membrane (PM), microbody (MB), and
cytosol (CS) (Agrawal et al., 2004). Three
Arabidopsis thaliana AOS genes (AtAOS1-3)
have been identified. All AtAOS genes are located
on chromosome 5. Arabidopsis thaliana AOS
proteins localize on chloroplast (CP), microbody
(MB), mitochondria (MC). In order to increase the
level of endogenous JA in rice, we try to over
express AOS genes by transforming AtAOS2 gene
to the rice genome.........................................
Fig. 1. The octadecanoid pathway leading to jasmonic acid (JA) biosynthesis in plants
AOC: allene oxide cyclase; OPR: 12-oxo-phytodienoic acid reductase; 13 (S)-HPOT: (9Z, 11E, 15Z,
13S)-13 - hydroperoxy - 9, 11, 15-octadecatrienoic acid; 12, 13 - EOT: (9Z, 11E, 15Z, 13S, 12R) - 13 -
Nguyen Thi Thuy Hanh, Han Oksoo
11
epoxy - 9, 11, 15 - octadecatrienoic acid; OPDA: 12 - oxo- 10, 15 (Z) - octadecatrienoic acid; OPC - 8:0:
3-oxo-2 (2' (Z) -pentenyl)-cyclopentane-1-octanoic acid (Schaller, 2001).
Fig. 2. Phylogenetic tree of AOS homologues and related proteins from other species
CP: chloroplast; CPS: chloroplast stroma; CS: cytosol; ER: endoplasmic reticulum; MB: microbody;
MC: mitochondria; PM: plasma membrane (Agrawal et al., 2004).

Application of PCR to identify the transgene (Arabidopsis thaliana allene oxide synthase2)...
12
Fig. 3. Structure of the pCAMBIA1201/AtAOS2 binary vector
(pCAMBIA1201 from CAMBIA,USA; pCAMBIA1201/AtAOS2 was contructed by Dr.Eunsun Kim)
In this study, a set of PCRs was carried out to
detect the hygromycin resistance gene, CaMV35S
promoter (the promoter located in front of the
AtAOS2 gene), and the AtAOS2 gene in the
transgenic rice genome. The results of PCRs would
confirm the appearance of the AtAOS2 gene in the
transgenic rice genome.
2. MATERIALS AND METHODS
2.1. Extraction of rice DNA
The leaves of one-month-old wild-type (Oryza
sativa L. Japonica cv. Nakdong) and 5 lines of T1
transgenic rice plants (T1 transgenic seeds were
obtained from Dr.Eunsun Kim) were harvested,
frozen in liquid nitrogen, and ground to fine
powder with a mortar and pestle under liquid
nitrogen. Total genomic DNA from non-transgenic
and transgenic rice plants were extracted by using
the DNeasy Plant Mini Kit (QIAGEN) according to
the instructions of the manufacturer.
2.2. PCR analysis
2.2.1. Design of Primers
The Primer 3 program was used to design all
primers for PCRs analysis.
2.2.2. PCR analysis to detect hygromycin
resistance gene
The primer pair employed to detect the
selectable marker (hygromycin
phosphotransferase) was: foward primer 5'-
ACAGCGTCTCCGACCTGATGCA-3' and
reverse primer 5'-
AGTCAATGACCGCTGTTATGCG-3'. PCR was
carried out according to the PCR Amplification
Kit by using HiPi PCR Premix (ELPis Biotech,
Korea) in a 20 µl reaction volume, with thermal
cycling parameters as follows: initial
denaturation at 940C for 5 min; following by 35
cycles of 940C for 1 min, 610C for 1 min, 720C
for 2 min; and 720C for 5 min for final extension.
2.2.3. PCR analysis to detect the CaMV
35S promoter
The primer pair, forward primer 5'-
GACCTAACAGAACTCGCCGTA-3' and
reverse primer 5'-
CACTTGCTTTGAAGACGTGGT-3', was
designed from the sequence of CaMV35S
promoter which located infront of AtAOS2 gene
in pCAMBIA1201/AtAOS binary vector (Fig. 3).
PCR was carried out according to the PCR
Amplification Kit by using HiPi PCR Premix
(ELPis Biotech, Korea) in a 20 µl reaction
volume. Template DNA were initially denatured
at 940C for 5 min, followed by 35 cycles with
thermal cycling parameters of 940C for 50 second,
580C for 50 seconds, 720C for 1 min.30 seconds.
A final 5 min. incubation at 720C was allowed for
completion of primer extension.
2.2.4. PCR analysis to detect the introduced
gene- AtAOS2
The primer pair employed to detect the
introduced genes was: forward primer 5'-
ACGACCAAGGAGCTGAAGAG-3' and reverse
primer 5'-CGCCGGTGGTAGACTAAATG-3'.
PCR was carried out according to the PCR
Amplification Kit by using HiPi PCR Premix
(ELPis Biotech, Korea) in a 20 µl reaction volume
with thermal cycling parameters as follows: 940C
for 5 min; following by 35 cycles of 940C for 40
seconds, 530C for 40 seconds, 720C for 1 min.40
seconds; and 720C for 5 min.
3. RESULTS AND DISCUSSION
The agarose gel banding patterns of the PCR
products were shown in figure 4. For the
hygromycin resistance gene, a 590 base pair
amplified segment was detected in
pCAMBIA1201/AtAOS plasmid and transgenic
rice genome with the specific designed primers,
but was not detected in non-transgenic (Fig. 4a).
For the CaMV35S promoter, which was located
infront of the AtAOS2 gene, the amplified band
size was 399 base pairs, and it was detected in
the plasmid, transgenic rice genome (Fig. 4b) but
was not detected in the non-transgenic genome.
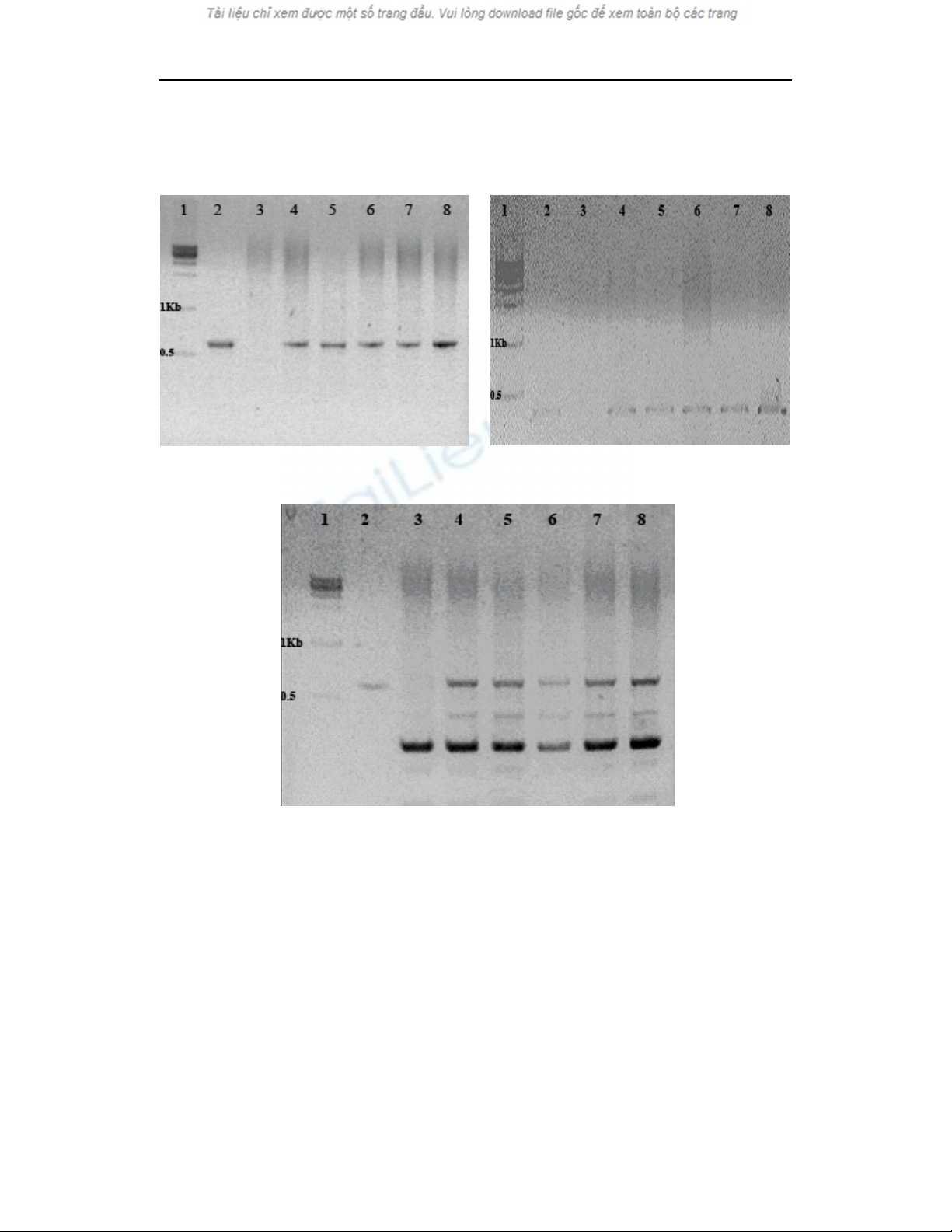
Nguyen Thi Thuy Hanh, Han Oksoo
13
A 599 base pair segment that was located in the
Arabidopsis thaliana cDNA region was detected
in the plasmid genome and transgenic rice
genome, but was not detected in the non-
transgenic rice genome (Fig. 4c).
fsghfjgykh
Fig. 4. Representative PCR analysis for the presence of the hygromycin resistance gene (a);
CaMV35S promoter (b) and AtAOS2 gene (c) in transgenic rice plants
Lane 1: molecular size marker; Lane 2:
pCAMBIA1201/AtAOS plasmid (positive control); Lane 3: untransformed plant (negative control);
Lanes 4 - 8: transformed plants.
















