
A unique variant of streptococcal group O-antigen (C-polysaccharide)
that lacks phosphocholine
Niklas Bergstro¨m
1
, Per-Erik Jansson
1
, Mogens Kilian
2
and Uffe B. Skov Sørensen
2
1
Clinical Research Centre, Analytical Unit, Karolinska Institute, Huddinge Hospital, Sweden;
2
Department of Medical Microbiology
and Immunology, University of Aarhus, Denmark
Streptococcus mitis strain SK598, which represents a sub-
group of biovar 1, possesses a unique variant of the
C-polysaccharide found in the cell wall of all strains of
Streptococcus pneumoniae and in some strains of S. mitis.
This new variant lacks the choline methyl groups in contrast
to the previously characterized forms of C-polysaccharide,
which all contain one or two choline residues per repeat. The
following structure of the repeating unit of the SK598
polysaccharide was established:
where AAT is 2-acetamido-4-amino-2,4,6-trideoxy-
D
-
galactose.
This structure is identical to the double choline-substi-
tuted form of C-polysaccharide, except that it is substituted
with ethanolamine instead of choline. This extends the
number of recognized C-polysaccharide variants to four.
Keywords: cell wall polysaccharide; C-polysaccharide; Strepto-
coccus pneumoniae; phosphocholine; Streptococcus mitis.
Previous serological analysis of the mitis group streptococci
suggested that C-polysaccharide is a common antigen of
Streptococcus pneumoniae and of most Streptococcus mitis
biovar 1 strains. Different reaction patterns, however,
emerged among the mitis group streptococci when exam-
ined by using a combination of two monoclonal antibodies
in an enzyme linked immunoassay that recognize phospho-
choline moieties and the backbone of C-polysaccharide,
respectively. Positive reactions with both monoclonals were
interpreted as the presence of the classical C-polysaccharide
with one or more phosphocholine residues attached, as
confirmed by structural analysis of polysaccharide prepared
from S. mitis strain SK137 [1]. Reaction with both of the
monoclonals was observed for all strains of S. pneumoniae
andforamajorityofS. mitis biovar 1 strains. However,
other strains reacted with one of the two monoclonals only,
and some S. mitis biovar 2 did not react with any of them.
The structure of the polysaccharide prepared from S. mitis
strain SK598, which represents strains that reacted with
the monoclonal antibody directed to the backbone of
C-polysaccharide but not with monoclonal antibody to
phosphocholine, is examined in the present study. It is
concluded that this S. mitis biovar 1 strain possesses a
unique variant of double choline-substituted C-polysaccha-
ride that lacks only the methyl groups in choline, i.e. is
substituted with ethanolamine residues. This new structural
variant extends the number of recognized C-polysaccharide
forms to four.
Materials and methods
Bacterial strain
The S. mitis biovar 1 strain SK598 used for preparation of
polysaccharide was from our own strain collection. This
strain was selected as it was negative for the presence of
phosphocholine, although it seemed to possess a C-poly-
saccharide like molecule when examined by ELISA and by
immunoelectrophoresis [1]. Strain SK598 was characterized
and identified as previously described [1,2]. It belongs to
Lancefield serogroup O as an extract from SK598 reacts
with streptococcal group O-antiserum purchased from
Statens Serum Institut, Copenhagen, Denmark.
Preparation of polysaccharide
The S. mitis biovar 1 strain SK598 was cultured overnight
at 37 C in 5 L laboratory flasks each containing 2.5 L
Todd-Hewitt broth (CM189, Oxoid, Basingstoke, UK).
The bacterial cells were harvested by centrifugation
Correspondence to P.-E. Jansson, Karolinska Institute,
Clinical Research Centre, Novum, Huddinge University Hospital,
S-141 86 Huddinge, Sweden.
Fax: + 46 8585 83820, Tel.: + 46 8585 83821,
E-mail: pererik.jansson@kfc.hs.sll.se
(Received 5 September 2002, revised 6 March 2003,
accepted 13 March 2003)
Eur. J. Biochem. 270, 2157–2162 (2003) FEBS 2003 doi:10.1046/j.1432-1033.2003.03569.x

(10 000 g, 30 min) and pooled from a total of 30 L broth
culture. The cells were washed twice in saline and suspended
in 50 mL of lysis buffer [0.1
M
NaCl, 1 m
M
MgCl
2
,0.05
M
Hepes pH 7.0, mutanolysin 100 UÆmL
)1
and lysozyme
1mgÆmL
)1
(M-9901 and L-6876, respectively, Sigma,
St Louis, MI, USA)]. Sodium azide (1 mgÆmL
)1
) was added
to the suspension as a preservative, and the bacterial cells
were digested at 37 C for 18 h. Cell debris was removed
from the digest by centrifugation and the supernatant was
heated to 50 C for 30 min to kill viable cells. Crude
polysaccharide was prepared by removal of most protein
and lipids from the lysate by chloroform/butanol treatment
followed by precipitation with ethanol [3]. The precipitate
was re-dissolved in MilliQ water, clarified by centrifugation
and lyophilized. The crude polysaccharide was treated with
DNAse, RNAse and proteinase K according to the manu-
facturer’s instructions and was then fractionated by size
exclusion chromatography on a Sephacryl S-300 column.
NMR spectroscopy
1
Hand
13
C NMR spectra were recorded with a JEOL JNM
ECP500 spectrometer, using standard pulse sequences.
Spectra of samples in 20 m
M
phosphate buffers of pD 7.4
were recorded at 35 C. Chemical shifts are reported
in p.p.m., using sodium 3-trimethylsilylpropanoate-d
4
(d
H
0.00) or acetone (d
C
31.00) or aqueous 2% phosphoric acid
(d
P
0.00) as internal references. For
13
Cand
31
P the reference
measurement was made with a separate tube before the
actual measurement. Chemical shifts were taken from 1D
spectra when possible, or else from
1
H,
1
H-correlated 2D
NMR spectra, i.e.
1
H,
1
H-COSY and
1
H,
1
H-TOCSY (40 ms
spin lock time). The mixing time in the NOESY experiment
was 300 ms. The J
H1,H2
values were obtained from the 1D
spectra, other couplings from the COSY spectrum. The
proton-carbon correlated spectrum (HMQC), and the
long-range proton-carbon correlated spectrum (HMBC)
were obtained with decoupling [4] using delay times of
42 and 97 ms using JEOL standard pulse sequences. The
delay time in the HMQC-TOCSY experiment was 20 ms.
The decoupled proton-phosphorus correlated spectra com-
prised a delay time of 71 ms, corresponding to 7 Hz
couplings.
Sugar and methylation analyses
For sugar analysis, alditol acetates were prepared by
hydrolysis of the polysaccharide using 2
M
trifluoroacetic
acid at 120 C for 2 h or 4
M
HCl at 120 Cfor1h,
followed by reduction with NaBH
4
or NaBD
4
,and
acetylation. For methylation analysis, methylation was
performed with methyl iodide in the presence of sodium
methyl sulfinyl methanide, and the methylated products
were purified using Sep-Pak C
18
-cartridges. For GLC, a
Hewlett-Packard 5890 instrument fitted with a flame-
ionization detector was used. Separation of alditol acetates
was performed on a DB-5 capillary column (30 m ·
0.25 mm) using a temperature program 160 C(1min)fi
250 Cat3CÆmin
)1
. GLC-MS (EI) was performed on
a Hewlett-Packard 5890/Nermag R10–10H quadrupole
instrument. Partially methylated alditol acetates were
separated on a DB-5 capillary column (25 m ·0.20 mm),
using the same temperature program as described for alditol
acetates. The absolute configurations of the sugar residues
were determined by GLC-MS of the trimethylsilylated
(+)-2-butyl glycosides [5], using the same temperature
program as described for alditol acetates.
HF degradation
A solution of the crude cell wall polysaccharide (69 mg) in
aqueous 48% HF (1 mL) was kept for 48 h at 18 C, blown
to dryness with dry air and residual traces of acid were
neutralized with 1
M
ammonia, and the resulting material
fractionated on a column of Bio-Gel P-4 eluted with 0.1
M
pyridinium acetate buffer at pH 5.3. Polymeric material
(minor) was recovered at the void volume and oligomeric
material at 1.4 void volumes (major).
Mass spectrometry
ESI-MS was performed in the negative mode using an LCQ
iontrap (Thermo Finnigan) with aqueous 50% acetonitrile
as the mobile phase at a flow rate of 10 lLÆmin
)1
.Samples
were dissolved in aqueous 50% acetonitrile at a concentra-
tion about 1 mgÆmL
)1
,and10lL was injected via a syringe
pump into the electrospray source.
Results
Size exclusion chromatography of the crude polysaccharide
from S. mitis SK598, pretreated to remove proteins, lipids
and nucleic acids, gave two partially overlapping peaks that
appeared at 1.3 (PSI) and 1.7 (PSII) void volumes in the
eluate from a Sephacryl S-300 column. The unseparated
material showed on hydrolysis with trifluoroacetic acid
ribitol, glucose, galactose, glucosamine and galactosamine
in the proportions, 1 : 1.8 : 1.4 : 1 : 0.2. PSI was a minor
fraction only (< 10%) and it was not investigated in detail
as it was a complex mixture of probably peptides and
polysaccharides. On trifluoroacetic acid hydrolysis it gave
ribitol, glucose, galactose in the ratio 1 : 3.5 : 3.5 and some
minor amounts of other monosaccharides.
The latter major fraction, PSII was hydrolyzed with 4
M
hydrochloric acid and showed glucose and galactosamine
in the proportions 1 : 4.5. This hydrolysis enhances amino
sugars but ribitol is not detected. The absolute configuration
of the sugars was
D
, as demonstrated by GLC of the
trimethylsilylated (+)-2-butyl glycosides. In order to main-
tain a constant pD to get reproducible spectra in the NMR
studies, the solution of PSII was buffered at pD 7.4
(pH 7.0). The
1
H-NMR spectrum of PSII showed five
major peaks in the anomeric region corresponding to
approximately one proton each, and some smaller signals
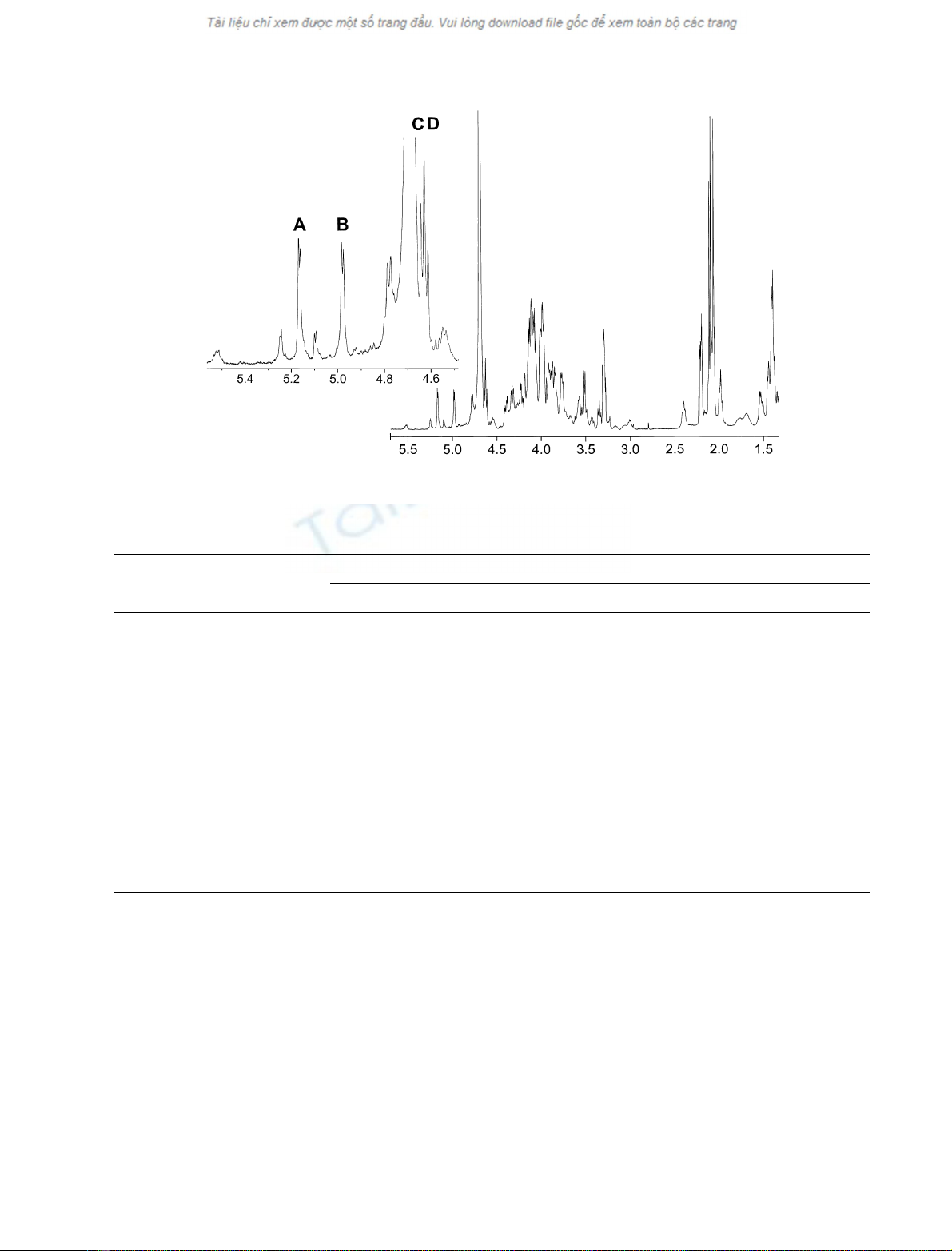
(Fig. 1). The five large signals in the anomeric region
appeared at d5.17, 4.94, 4.77, 4.64 and 4.62 (Table 1). This
could be recognized as closely similar but not identical to
signals in the anomeric region from the C-polysaccharide
purified from S. pneumoniae [1,3,6–8]. Four of the signals
could be shown to be anomeric and appeared at d5.17 (J
1,2
3.5 Hz, 1H), 4.94 (J
1,2
3.5 Hz, 1H), 4.64 (J
1,2
7.3 Hz, 1H),
and 4.62 (J
1,2
7.3 Hz, 1H) and the corresponding sugar
residues were designated A–D, respectively. A signal at
d4.77, which was an obscured quartet, could be assigned to
2158 N. Bergstro
¨met al. (Eur. J. Biochem. 270)FEBS 2003
H-5 of a 2-acetamido-4-amino-2,4,6-trideoxy-
D
-galactose
residue (AAT) (see below).
Asignalatd3.29–3.30 (4 H) was assigned to two
N-linked methylene groups in two phosphoethanolamine
residues (see below). Four signals for anomeric carbons,
virtually coinciding with those reported previously for the
C-polysaccharide [1,3,6–8], were observed in the
13
C-NMR
spectrum at d104.6, 102.1, 98.9, and 94.2.
For residues Aand Dit was possible to follow the spin-
systems from H-1 up to H-4 in the COSY spectrum. For
residues Band Cit was possible to follow the whole spin-
system in the COSY spectrum, these assignments were then
verified in the TOCSY spectrum. Residue A(H-1 d5.17)
could be assigned to a 4,6-disubstituted GalNAc residue
with the aconfiguration, as evident from its J
1,2
-value of
3.5 Hz. The galacto configuration was apparent as the H-3–
H-4 coupling was small. That C-2 was linked to nitrogen
was indicated by a correlation in the HMQC spectrum to a
signal at d50.1. The C-5 signal was identified from a
correlation from H-1 in the HMBC spectrum. H-5 and H-6
were both identified by a correlation to C-4 in the HMBC
spectrum; correlations between H-5/C-6 and H-6/C-5
verified the assignments. Substituted positions in the residue
were indicated from the large glycosylation shifts, 7.8 and
1.9 p.p.m., for the C-4 and C-6 signals, respectively, when
compared to unsubstituted a-
D
-GalNAc. Residue B(H-1
Fig. 1.
1
H NMR spectrum (35 °C, 500 MHz) of the cell wall polysaccharide from S. mitis SK598. A–D refer to anomeric protons as described in the
text.
Table 1.
1
H- and
13
C-NMR data for the C-polysaccharide (PSII) of S. mitis.SK598 obtained at pD 7.4.
Sugar residue
Chemical shifts (p.p.m.)
1 2345 6a6b
fi6)-a-GalpNAc(1fiA5.17 [3,5]
a
4.32 3.93 4.11 4.01 4.02 4.02
4 94.2 50.1 67.5 77.4 71.3 64.0
›
fi3)-a-AATp(1fiB4.98 [3,5] 4.23 4.39 3.94 4.77 1.24
98.9 49.0 75.6 55.3 63.7 16.0
fi6)-b-Glcp-(1fiC4.64 [3,7] 3.35 3.51 3.52 3.57 4.10 4.14
104.6 73.5 76.0 69.4 75.1 65.0
fi6)-b-GalpNAc(1fiD4.62 [3,7] 4.11 3.86 4.18 3.84 4.07 4.07
3 102.1 51.1 75.0 63.9 74.0 65.0
›
fi1)-Ribitol(5fiE3.89, 3.99 3.77 3.91 3.77 3.98, 4.07
71.3 72.2 71.4 72.2 67.0
Ethanolamine F4.09 3.29
62.5 40.7
Ethanolamine G4.13 3.30
62.5 40.7
a
J
1,2
-values are given in brackets.
FEBS 2003 Cell wall polysaccharides of S. mitis biovar 1 (Eur. J. Biochem. 270) 2159


























