
A functional role of the membrane-proximal extracellular domains
of the signal transducer gp130 in heterodimerization
with the leukemia inhibitory factor receptor
Andreas Timmermann, Andrea Ku¨ ster, Ingo Kurth, Peter C. Heinrich and Gerhard Mu¨ ller-Newen
Institut fu
¨r Biochemie, Rheinisch-Westfa
¨lische Technische Hochschule Aachen, Germany
gp130 is the common signal transducing receptor subunit of
interleukin (IL)-6-type cytokines. gp130 either homodimer-
izes in response to IL-6 and IL-11 or forms heterodimers
with the leukemia inhibitory factor (LIF) receptor (LIFR) in
response to LIF, oncostatin M (OSM), ciliary neurotrophic
factor (CNTF), cardiotrophin-1 (CT-1) or cardiotrophin-
like cytokine resulting in the onset of cytoplasmic tyrosine
phosphorylation cascades. The extracellular parts of both
gp130 and LIFR consist of several Ig-like and fibronectin
type III-like domains. The role of the membrane-distal
domains of gp130 (D1, D2, D3) and LIFR in ligand binding
is well established. In this study we investigated the func-
tional significance of the membrane-proximal domains of
gp130 (D4, D5, D6) in respect to heterodimerization with
LIFR. Deletion of each of the membrane-proximal domains
of gp130 (D4, D5andD6) leads to LIF unresponsiveness.
Replacement of the gp130 domains by the corresponding
domains of the related GCSF receptor either restores weak
LIF responsiveness (D4-GCSFR), leads to constitutive
activation of gp130 (D5-GCSFR) or results in an inactive
receptor (D6-GCSFR). Mutation of a specific cysteine in D5
of gp130 (C458A) leads to constitutive heterodimerization
with the LIFR and increased sensitivity towards LIF
stimulation. Based on these findings, a functional model of
the gp130–LIFR heterodimer is proposed that includes
contacts between D5 of gp130 and the corresponding
domain D7 of the LIFR and highlights the requirement for
both receptor dimerization and adequate receptor orienta-
tion as a prerequisite for signal transduction.
Keywords: cytokines; receptors; signal transduction; leuke-
mia inhibitory factor; gp130.
Secretion of mediators by cells that are recognized by
specific receptors on target cells is a basic mechanism of
intercellular communication. The molecular mechanism by
which binding of the ligand to the receptor on the plasma
membrane leads to the onset of cytoplasmic signal trans-
duction cascades has gained considerable attention during
recent years. In the case of receptors that span the
membrane only once, ligand induced receptor dimerization
has been accepted as the main mechanism for receptor
activation [1]. Only recently, several reports suggested that
some receptors may exist as preformed dimers or multimers
that switch from an inactive to an active conformation upon
ligand binding [2,3].
Hematopoietic cytokine receptors [4] consist of an
extracellular part, a single transmembrane region, and a
cytoplasmic part that is devoid of any intrinsic enzymatic
activity but constitutively associates with tyrosine kinases of
the Janus kinase (Jak) family. Upon ligand binding the
associated Jaks become activated by transphosphorylation
and phosphorylate tyrosine residues in the cytoplasmic part
of the receptor. These phosphotyrosines serve as docking
sites for signalling molecules that, in most cases, also
become phosphorylated. Most importantly, STAT (signal
transducer and activator of transcription) factors are
recruited to the receptor, dimerize upon phosphorylation
and translocate into the nucleus to induce expression of
target genes [5].
Based on the architecture of the extracellular part,
hematopoietic cytokine receptors can be subdivided into
two groups. The extracellular parts of short cytokine
receptors like erythropoetin recepter (EpoR), growth
hormone receptor (GHR), prolactinR, IL-2Rbor IL-4R
consist of only a single cytokine binding module (CBM).
The CBM is made up of two fibronectin type III-like
(FNIII) domains containing some characteristic conserved
motifs in their primary structures. Several structures of
CBMs of short cytokine receptors bound to their ligands
have been solved showing that in the active receptor dimer
the membrane-proximal domains are juxtaposed in a well-
defined orientation [6,7].
The extracellular parts of complex cytokine receptors like
gp130, LIFR, leptinR or GCSFR contain at least one CBM
and additional FNIII- and Ig-like domains. The cytokine
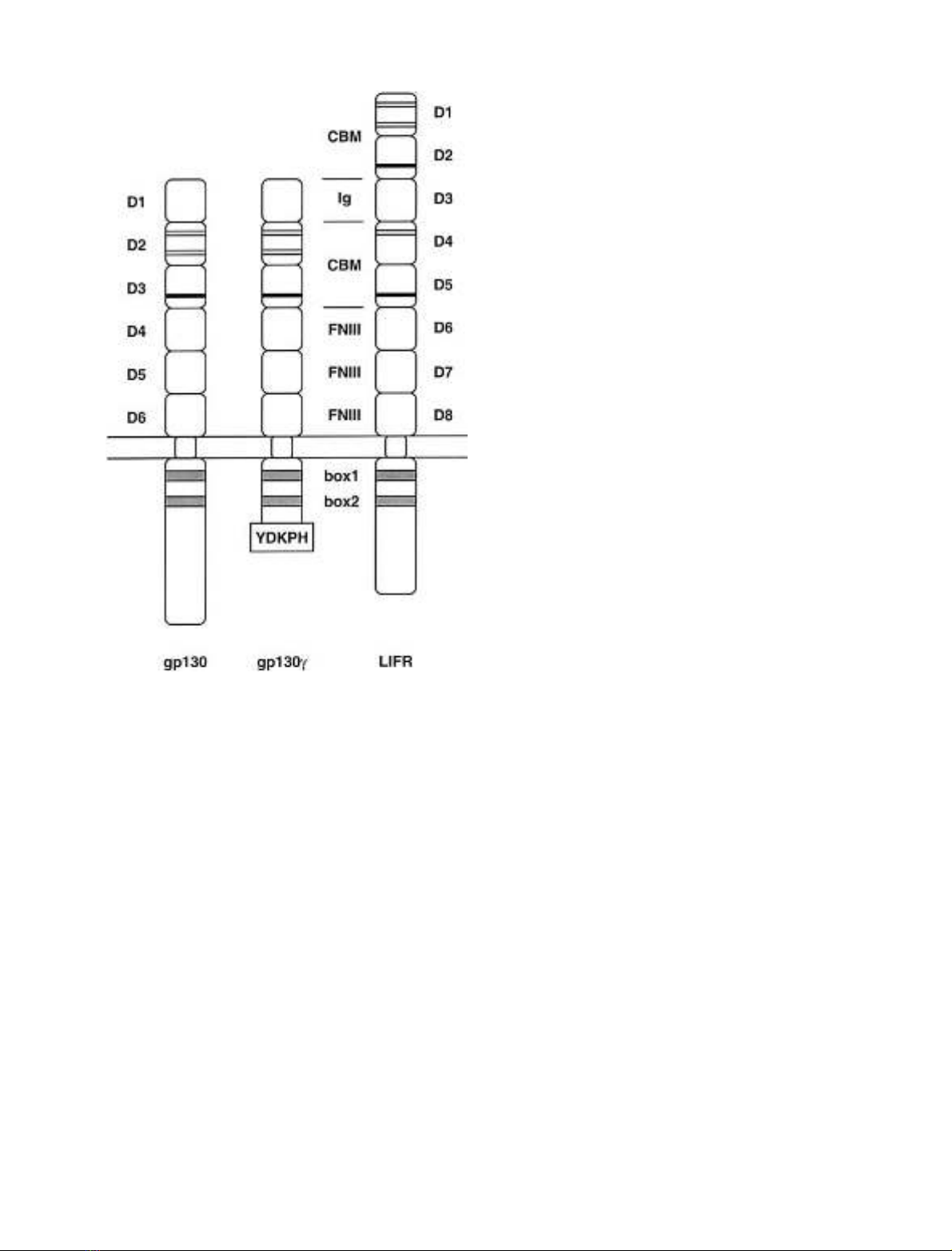
receptor gp130 consists of an Ig-like domain (D1), followed
by a CBM (D2, D3) and three FNIII-like domains (D4, D5,
and D6) (Fig. 1) [8]. The role of the membrane-distal
domains (D1–D3) in ligand binding has been well estab-
lished by functional and structural studies. In response
Correspondence to G. Mu
¨ller-Newen, Institut fu
¨r Biochemie,
Rheinisch-Westfa
¨lische Technische Hochschule Aachen,
Pauwelsstr. 30, D-52057 Aachen, Germany.
Fax: + 49 241 8082428, Tel.: + 49 241 8088860,
E-mail: Mueller-Newen@RWTH-Aachen.de
Abbreviations: CBM, cytokine binding module; FNIII, fibronectin
type III-like; GCSF, granulocyte colony stimulating factor; GH,
growth hormone; IL, interleukin; Jak, Janus kinase; LIF, leukemia
inhibitory factor; OSM, oncostatin M; STAT, signal transducer and
activator of transcription.
Note: a web site is available at http://www.biochem.rwth-aachen.de
(Received 28 February 2002, accepted 18 April 2002)
Eur. J. Biochem. 269, 2716–2726 (2002) FEBS 2002 doi:10.1046/j.1432-1033.2002.02941.x
to cytokines like IL-6 and IL-11 that lead to gp130
homodimerization, two different epitopes of gp130 are
involved in ligand binding: the Ig-like domain and the CBM
[9–11]. In the case of LIF-induced heterodimerization of
LIFR with gp130, the cytokine first binds to the Ig-like
domain of the LIFR [12,13]. gp130 is recruited to the LIF–
LIFR complex via its CBM without involvement of its
Ig-like domain [14,15]. The cytokine oncostatin M (OSM)
first binds to gp130 and then induces heterodimerization
with LIFR [12] or OSMR [16].
Besides the CBM and Ig-like domains both gp130 and
LIFR share three further membrane-proximal FNIII-like
domains as a common structural feature [17]. In a previous
study we established a functional role for each of the
membrane-proximal domains of gp130 for receptor activa-
tion in response to ligands that lead to gp130 homodime-
rization. We proposed a particular role for D5 of gp130 in
respect to proper receptor spacing and orientation [18]. In
this study, using gp130 deletion mutants, point mutants and
chimeric receptors, the role of the individual membrane-
proximal domains of gp130 in heterodimerization with the
LIFR is evaluated. A cysteine to alanine mutation in D5 of
gp130 in combination with the LIFR leads to a weak
constitutive activity and an elevated response to stimulation
with LIF. A model for the gp130/LIFR interaction is
proposed, in which D5 of gp130 contacts domain 7 of the
LIFR.
MATERIALS AND METHODS
Enzymes, proteins, antibodies, chemicals,
and cell culture media
Enzymes were purchased from Roche (Mannheim, Ger-
many) and protein A–Sepharose was obtained from
Amersham (Freiburg, Germany). Fugene was obtained
from Roche (Mannheim, Germany). DMEM and antibi-
otics were obtained from Life Technologies (Eggenstein,
Germany); fetal bovine serum was provided by Seromed
(Berlin, Germany). [a-
32
P]deoxyATP was purchased from
Hartmann Analytic (Braunschweig, Germany). Human
rIL-6 was expressed in Escherichia coli, refolded, and
purified as described by Arcone et al.[19].Thespecific
activity was 10
8
units per mg of protein in the B9 cell
proliferation assay [20]. Soluble IL-6R (sIL-6R) [21] was
expressed in insect cells as described previously. The gp130
mAbs B-P4 and B-P8 and the LIFR mAb 10B2 were
generated as described elsewhere [22,23]. The polyclonal
LIFR antiserum sc-659 was obtainded from New England
Biolabs (Frankfurt/Main, Germany). All other Abs were
purchased from Dako (Hamburg, Germany). NaCl/P
i
buffer contained 200 m
M
NaCl, 2.5 m
M
KCl, 8 m
M
Na
2
HPO
4
,and1.5m
M
KH
2
PO
4
.
Cell culture
BaF3-cells, a murine pro-B lymphocyte line, were cultured in
RPMI 1640 containing 10% fetal bovine serum, 100 mgÆL
)1
streptomycin, 60 mgÆL
)1
penicillin and 5% conditioned
medium from X63Ag-653 BPV-mIL-3 myeloma cells as a
source of IL-3. Simian monkey kidney cells (COS7) were
cultured in DMEM supplemented with 10% fetal bovine
serum, 100 mgÆL
)1
streptomycin, and 60 mgÆL
)1
penicillin.
Cells were grown at 37 C in a water-saturated atmo-
sphere at 5% CO
2
. BaF3 transfectants were cultured in the
presence of 0.5 lgÆmL
)1
hygromycin if transfected with the
LIFR expression vector pSBC1/2-LIFR/Hygro and
1mgÆmL
)1
G418 if transfected with a pSVL-gp130-expres-
sion vector together with pSV2-Neo.
All cells were regularly checked for the absence of
mycoplasma infection using PCR detection of mycoplasma
DNA.
Plasmid construction
Construction of gp130 wild-type and domain mutant
expression vectors D4, D5, D6 and D5 has been described
elsewhere [18]. The domain exchange mutants gp130 D4
and D6 were cloned analogously after amplifying the DNA
Fig. 1. Schematic representation of gp130, gp130cand LIFR. The
predicted structural organizations of gp130, gp130cand LIFR are
shown. Black lines in the CBM indicate conserved cysteine residues,
black bars the WSXWS motifs. The Ig-like domains and the mem-
brane-proximal FNIII domains are labelled. The extracellular domains
of gp130 and LIFR are numbered from domain 1 (D1) to domain 6
(D6) or domain 1 (D1) to domain 8 (D8), respectively. In the cyto-
plasmic part of gp130c, the amino-acid residues following the Jak
binding sites (box1 and box2, gray boxes) were replaced by the strongly
and specifically STAT1-activating motif YDKPH of the interferon-c
receptor.
FEBS 2002 Heterodimerization of gp130 with LIFR (Eur. J. Biochem. 269) 2717

encoding domains 4 and 6 of GCSFR using the oligo-
nucleotides: 5¢-ACTACCGAACGGGCCCCCGGGGTC
AGACTGGACACATGG-3¢and 5¢-TCGGGCCATGGC
ATGCCCGGGGGTCAGAGCTGGG-3¢for amplifica-
tion of D4 of GCSFR and 5¢-TACTCTCAAGAAATG
CCCGGGTCCCATGCCCCAGAG-3¢and 5¢-GCCCAG
GATGATGTGTAGCTCCCCGGGCTCTGGGGTCAA
GGT-3¢for D6 of GCSFR (the XmaI sites are underlined)
as PCR primers. Starting point for cloning of gp130 C458A,
C466A and C491A was the full length human gp130 cDNA
cloned into the XhoIandBamHI site of the eukaryotic
expression vector pSVL lacking the EcoRI site (gp130-
pSVLDEco). Using this vector as template, for each point
mutant two fragments were amplified. In a first reaction, the
DNA was amplified using the primer pSVL(sense) and an
antisense primer containing the mutation. A second PCR-
fragment was generated using the primers pSVL(antisense)
and a sense primer with the corresponding mutation. These
fragments were isolated, mixed and served as templates for a
fusion PCR using the primers pSVL(sense) and pSVL(anti-
sense). The reaction products were digested with the
restriction enzymes Xho IandBstEII and cloned into the
expression vector gp130-pSVLDEco. The primers used for
the PCR reactions were: pSVL(sense) 5¢-GTGTTACTT
CTGCTCT-3¢; pSVL(antisense) 5¢-TCTAGTTGTGGTT
TGT-3¢; C458A(sense) 5¢-ATACTTGAGTGGGCTGTG
TTATCAG-3¢; C458A(antisense) 5¢-ATCTGATAACAC
AGCCCACTCAAGTAT-3¢; C466A(sense) 5¢-GATAAA
GCACCCGCTATCACAGACTGG-3¢; C466A(antisense)
5¢-CCAGTCTGTGATAGCGGGTGCTTTATCTG-3¢;
C491A(sense) 5¢-GCAGAGAGCAAAGCCTATTTGAT
AACAG-3¢and C491(antisense) 5¢-TGTTATCAAATAG
GCTTTGCTCTCTG-3¢.
PCRs were performed applying standard procedures. All
plasmids were sequenced using an ABI Prism Automated
sequencer (Applied Biosystems).
The full-length human LIFR cDNA was cloned into
pSBC-1 to yield the mammalian expression vector pSBC-
LIFR as previously described [15]. For the transfection of
BaF3-cells, the bicistronic expression vector pSBC1/2-
LIFR/Hygro was used [15,24].
Transfection of cells
Plasmid DNA was transfected into BaF3-cells by electro-
poration. Thirty micrograms of the bicistronic LIFR
expression vector pSBC1/2-LIFR-Hygro were electropo-
rated into 3.5 ·10
6
cells in 0.8 mL medium applying a
single 70-ms pulse at 200 V. Selection with hygromycin
(0.5 mgÆmL
)1
) was initiated 24 h after transfection. Selected
BaF3 clones were screened for the presence of membrane-
bound LIFR proteins by flow cytometry. For transfection
of gp130 constructs, 28 lg of the gp130 expression vector
were coelectroporated with 2 lg of pSV2neo as described
above. Selection with G418 (3 mgÆmL
)1
) was initiated 24 h
after transfection. For transfection, either untransfected
BaF3-cells or cells previously transfected with pSBC1/2-
LIFR-Hygro were used. Selected BaF3 clones were screened
for the presence of membrane-bound gp130 proteins by
flow cytometry.
COS7 cells were transiently transfected using the Fugene
method. Efficiency of transfection was analysed by flow
cytometry.
Flow cytometry
Cells were collected, washed and resuspended in cold NaCl/
P
i
containing 5% fetal bovine serum and 0.1% sodium
azide. Subsequently, cells were incubated on ice with
10 lgÆmL
)1
gp130antibodiesB-P4orB-P8or10lgÆmL
)1
LIFR antibody 10B2. Cells were washed with cold NaCl/P
i
/
azide and incubated with R-phycoerythrin-conjugated anti-
(mouse IgG) Fab-fragment at a 1 : 50 dilution. Again, cells
were washed with cold NaCl/P
i
/azide and then resuspended
in 400 lLNaCl/P
i
/azide followed by flow cytometry
analysis using a FACScalibur (Beckton Dickinson).
Electrophoretic mobility shift assay (EMSA)
Cells were incubated at 37 C for 15 min in the presence of
IL-6/sIL-6R, LIF, OSM or left unstimulated. BaF3-cells
were stimulated with 25 ngÆmL
)1
IL-6 and 1 lgÆmL
)1
sIL-
6R or 50 ngÆmL
)1
LIF or 50 ngÆmL
)1
OSM. COS7 cells
were stimulated with 12.5 ngÆmL
)1
IL-6 and 500 ngÆmL
)1
sIL-6R, 20 ngÆmL
)1
LIF and 4 ngÆmL
)1
OSM. Where
indicated, cells were preincubated for 2 h in the presence of
500 l
M
2-mercaptoethanol prior to stimulation. Prepar-
ation of nuclear extracts and EMSAs were performed as
described previously [25]. A double stranded size-inducible
element (SIE) oligonucleotide derived from the c-fos
promoter (m67SIE; 5¢-GATCCGGGAGGGATTTACGG
GGAAATGCTG-3¢) was used as
32
P-labeled probe [26].
The protein–DNA complexes were separated on a 4.5%
polyacrylamide gel containing 7.5% glycerol. The electro-
phoresis was performed using 0.25 ·NaCl/Tris/borate
buffer at 20 VÆcm
)1
.
Coimmunoprecipitation of LIFR/gp130 complexes
Transiently transfected COS7 cells were stimulated for
15 min with IL-6/sIL-6R, LIF or OSM as described or left
unstimulated. Where indicated, cells were preincubated for
2 h in the presence of 500 l
M
2-mercaptoethanol prior to
stimulation. Immediately after stimulation, cells were washed
twice with ice-cold NaCl/P
i
containing 100 l
M
vanadate.
After addition of 600 lL lysis buffer (10% glycerol, 0.25%
Brij-96, 50 m
M
Tris/HCl, 50 l
M
Na
3
VO
4
,100l
M
EDTA,
1m
M
phenylmethanesulfonyl fluoride, 1 mgÆL
)1
aprotinin,
1mgÆL
)1
leupeptin, pH 8.0) the cells were collected and lysed
for 30 min in a microcentrifuge tube. The lysate was
centrifuged for 1 min at 3000 r.p.m. in an Eppendorf
centrifuge and the supernatant was transferred into a new
centrifuge tube. Following incubation of the lysate with
1.6 lg sc-659 antiserum for 12 h at 4 C15mgproteinA–
sepharose was added. After incubation for 12 h at 4 C, the
complexes were washed twice with NaCl/Tris/borate/Non-
idet P40 buffer, resuspended in Laemmli-buffer, incubated at
95 C for 5 min and separated on a 7% SDS polyacrylamide
gel under reducing conditions followed by electroblotting.
Immunoblotting and enhanced
chemiluminescence (ECL) detection
Immunoprecipitated proteins separated by SDS/PAGE
were transferred to a poly(vinylidene difluoride) membrane
by a semidry electroblotting procedure [27]. Poly(vinylidene
difluoride) membranes were blocked in a solution of 20 m
M
2718 A. Timmermann et al. (Eur. J. Biochem. 269)FEBS 2002

Tris/HCl (pH 7.6), 137 m
M
NaCl, 0.1% Nonidet-P40
containing 10% bovine serum albumin and probed with
antibody, followed by incubation with horseradish peroxi-
dase conjugated secondary antibody. Immunoreactive pro-
teins were detected by chemiluminescence using the ECL-kit
(Amersham, UK) following the manufacturer’s instruc-
tions.
RESULTS AND DISCUSSION
Each of the three membrane-proximal domains
of gp130 is required for signal transduction
in response to LIF and OSM
To investigate the role of the membrane-proximal FNIII-
domains of gp130 in signal transduction through hetero-
dimeric complexes with the LIFR, mutants of gp130, in
which single FNIII-domains are deleted were generated
lacking either D4 (gp130-D4), D5 (gp130-D5) or D6 (gp130-
D6) [18]. These gp130 mutants were coexpressed with the
LIFR in different cell types. The STAT-activation after
stimulation with the cytokines IL-6, LIF or OSM was used
as a measure of signal transduction through the analysed
complexes.
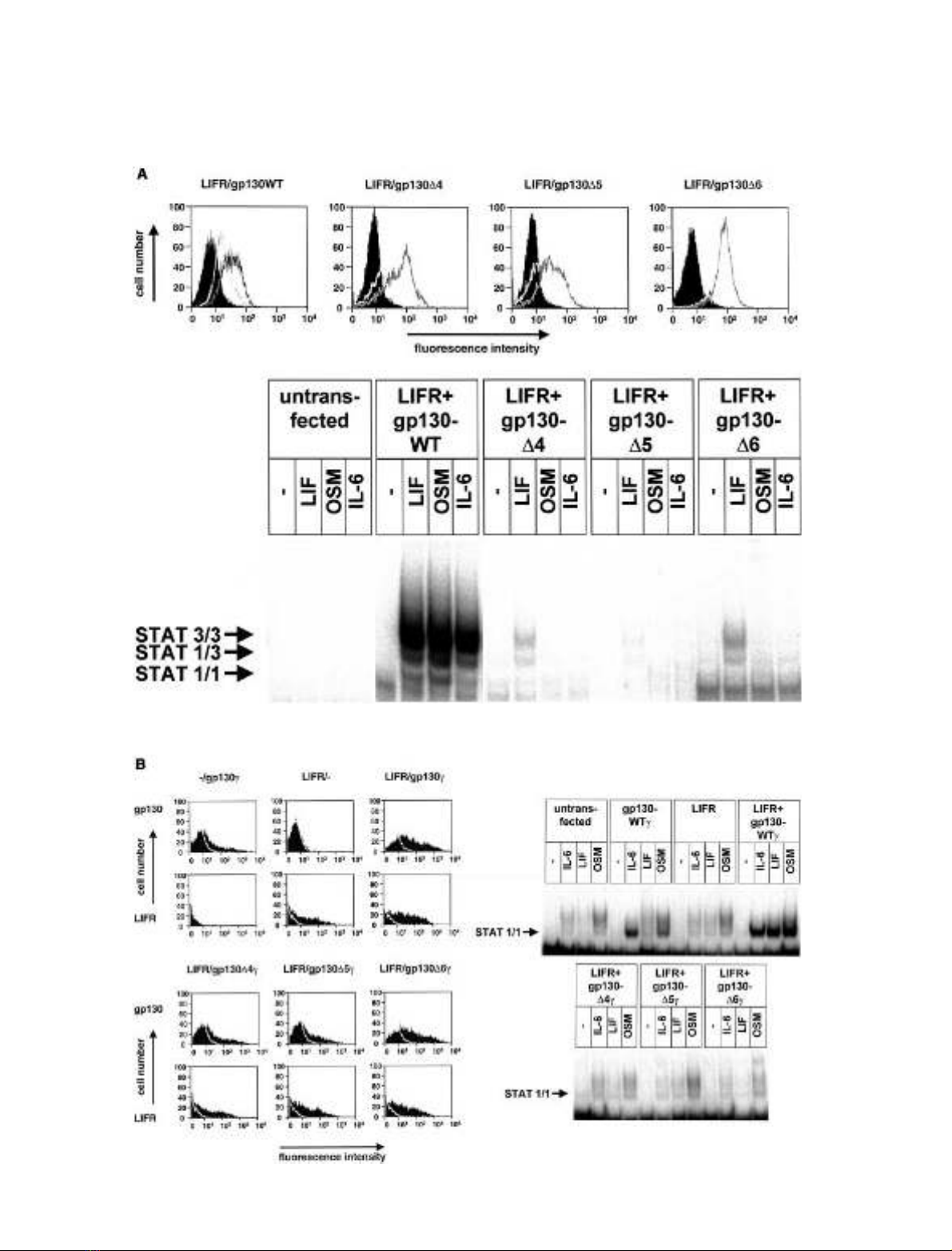
Cells of the murine pre-B cell line BaF3 do not express
endogenous gp130 or LIFR. After stably transfecting these
cells with the respective cDNAs, cell surface expression of
both receptors was detected. After stable transfection of the
deletion constructs gp130D4, gp130D5orgp130D6 together
with the LIFR expression vector in BaF3-cells, the surface
expressions of both receptors were similar to those detected
for wild-type gp130/LIFR transfected cells (Fig. 2A, upper
panel).
After stimulation of these cells with IL-6/sIL-6R, none of
the analysed mutants showed a STAT activation similar to
wild-type receptors (Fig. 2A, lower panel, right). This
confirms the previously reported inactivity of the deletion
mutants in response to the gp130-homodimerizing cytokine
IL-6 [18]. Interestingly, also the formation of active hetero-
dimers with wild-type LIFR in response to LIF or OSM is
strongly reduced or abolished by deletion of individual
membrane-proximal domains of gp130. Thus, in BaF3-cells,
each of the membrane-proximal domains of gp130 is
necessary for the efficient formation of a signal transducing
heterodimeric complex of gp130 and the LIFR.
To ensure that the measured receptor activation after
cytokine stimulation does not depend on the analysed
cellular environment, the deletion mutants were expressed
together with the LIFR in COS7 cells. In a previous report
[15], we established a system that allows the study of gp130
mutants together with the LIFR in COS7 cells despite the
presence of low amounts of endogenous wild-type receptors.
To achieve this, the cytoplasmic tyrosine motifs of gp130
that predominantly recruit STAT3 were replaced by the
STAT1 recruiting motif of the interferon-creceptor result-
ing in a chimeric protein designated gp130c.Inorderto
investigate the role of the membrane-proximal domains of
gp130 in receptor activation, the FNIII-domain deletions
were introduced into the gp130c-construct (gp130-D4c,
gp130-D5cor gp130-D6c) [18]. Each of these constructs was
cotransfected with the LIFR into COS7 cells. Enhanced
receptor surface expression was detected by flow cytometry
(Fig. 2B, left panel).
Transfected cells were stimulated with IL-6/sIL-6R, LIF
or OSM and subsequently activation of STAT1 was
analysed by EMSA (Fig. 2B, right panel). In mock-trans-
fected cells only a weak response to IL-6/sIL-6R and OSM
was observed due to endogenously expressed receptors
resulting in a low-level activation of mainly STAT3. Cells
transfected with gp130cshowed an increased STAT1
response to IL-6, and less pronounced to OSM. The LIF
response remained unchanged. Transfection of LIFR alone
did not significantly change the sensitivity of the cells to any
of the cytokines. Transfection of both gp130cand LIFR led
to a prominent response of the cells to all three cytokines.
No STAT1-activation after cytokine stimulation could be
detected when one of the gp130cdeletion constructs was
coexpressed with the LIFR. In COS7 cells under conditions
of receptor overexpression, as in BaF3-cells, each of the
membrane-proximal domains of gp130 is necessary for the
formation of signal transducing heterodimeric complexes of
gp130 and LIFR.
Two explanations for this finding can be discussed. The
first is based on the identical domain architecture of LIFR
and gp130 in the membrane-proximal six domains. This is
likely to result in the same distance between the cell surface
and the ligand-binding epitopes of both receptors. Deletion
of a single domain in the membrane-proximal part of gp130
leads to a shift of the receptor areas involved in ligand
binding closer to the membrane, resulting in the inability of
the receptor chains to form an active receptor dimer.
Additionally, the membrane-proximal domains can act as
contact sites between the signal transducing receptor chains
or can permit the signal competent conformation of gp130
homo- or heterodimers by adjusting a defined position
towards each other. Thus, deletion of a membrane-proximal
domain of gp130 may be without consequence on ligand
binding but lead to a larger distance or a twist of the
cytoplasmic parts of the receptors responsible for signal
transduction.
Replacement of single membrane-proximal
FNIII-domains of gp130 by corresponding domains of
GCSFR leads to different effects on signal transduction
To investigate, if the function of the membrane-proximal
domains of gp130 is limited to ensure the correct spacing
between the CBM and the membrane, each of the domains
was replaced by the corresponding domain of the GCSFR.
The replacement is assumed to compensate for the shift of
the ligand-binding epitopes of the receptors. The domain
architecture of GCSFR is identical to that of gp130;
moreover these receptors share 46% sequence homology.
The gp130 constructs with exchanged individual FNIII-
domains were introduced into the gp130c-construct result-
ing in the mutants gp130D4cand gp130D6c, respectively.
The construction of the corresponding mutant gp130D5c
has been previously described [18].
Upon co-expression of the gp130 chimeras with the LIFR
in COS7 cells, both receptors were expressed on the cell
surface as detected by flow cytometry in amounts similar to
those of gp130 wild-type (data not shown). Signal trans-
duction was measured by STAT1 activation in an EMSA
(Fig. 3). The exchange of individual membrane-proximal
FNIII-domains of gp130 by the corresponding domains of
the GCSFR resulted in a complex signal transduction
FEBS 2002 Heterodimerization of gp130 with LIFR (Eur. J. Biochem. 269) 2719
pattern. Cells transfected with gp130D5ctogether with the
LIFR showed a prominent STAT1-activation independ-
ently of stimulation. This activation was not enhanced by
stimulation with cytokines. These results are in line with
observations previously made in COS7 cells transfected
with the gp130D5c-construct alone [18]. When coexpressed
2720 A. Timmermann et al. (Eur. J. Biochem. 269)FEBS 2002


























