
Caspase-2 is resistant to inhibition by inhibitor of
apoptosis proteins (IAPs) and can activate caspase-7
Po-ki Ho
1,2,3
, Anissa M. Jabbour
1,2,3
, Paul G. Ekert
1,4,5
and Christine J. Hawkins
1,2,3
1 Murdoch Children’s Research Institute, Parkville, Australia
2 Children’s Cancer Centre, Royal Children’s Hospital, Parkville, Australia
3 Department of Paediatrics, University of Melbourne, Parkville, Australia
4 Department of Neonatology, Royal Children’s Hospital, Parkville, Australia
5 The Walter and Eliza Hall Institute, Royal Melbourne Hospital, Parkville, Australia
The caspases are a family of cysteine proteases that
typically cleave their substrates at aspartate residues
[1]. Subclassification of family members has been based
on various criteria including substrate specificity or
structural features. For example, caspases-1, -4 and -5
are involved in the proteolytic maturation of cytokines
including pro-interleukin-1b[2] and pro-interleukin-18
[3]. Caspases-8 and -9 are components of cell death
signal transduction pathways and are classified as api-
cal caspases. The primary role of these proteases, each
of which has a long prodomain containing a protein
interaction motif, is to proteolytically activate distal
caspases (such as caspase-3 and caspase-7), which then
catalyse the cleavage of numerous cellular substrates
[4]. Despite being the second identified member of the
caspase family, the function of caspase-2 (Nedd-2 ⁄
Ich-1) remains somewhat elusive. Its substrate prefer-
ence more closely aligns with that of the pro-apoptotic
caspases than their cytokine processing relatives [5]. Of
the mammalian caspases, caspase-2 is the most similar
to the nematode apoptotic caspase, CED-3. This
would also tend to imply that caspase-2 plays a pro-
apoptotic role, yet caspase-2 deficient mice have
an extremely subtle phenotype, arguing against a non-
redundant role in programmed cell death [6,7].
Caspase-2 has recently received considerable
attention, as several groups have sought to define its
biological role in apoptosis signalling. Overexpressing
caspase-2 provoked the release of pro-apoptotic mole-
cules (including cytochrome c) from mitochondria [8],
Keywords
caspase-2; protease; caspase-7;
S. cerevisiae; enzyme activity
Correspondence
C. Hawkins or P. Ekert, Murdoch Children’s
Research Institute, Royal Children’s
Hospital, Flemington Road, Parkville, VIC
3052 Australia
Fax: +61 3 9345 4993 (CH); +61 3 9347
0852 (PE)
Tel: +61 3 9345 5823 (CH); +61 3 9345
2548 (PE)
E-mail: chris.hawkins@mcri.edu.au;
paul.ekert@mcri.edu.au
(Received 10 November 2004, revised 7
January 2005, accepted 18 January 2005)
doi:10.1111/j.1742-4658.2005.04573.x
Caspases are a family of cysteine proteases with roles in cytokine matur-
ation or apoptosis. Caspase-2 was the first pro-apoptotic caspase identified,
but its functions in apoptotic signal transduction are still being elucidated.
This study examined the regulation of the activity of caspase-2 using
recombinant proteins and a yeast-based system. Our data suggest that for
human caspase-2 to be active its large and small subunits must be separ-
ated. For maximal activity its prodomain must also be removed. Consistent
with its proposed identity as an upstream caspase, caspase-2 could provoke
the activation of caspase-7. Caspase-2 was not subject to inhibition by
members of the IAP family of apoptosis inhibitors.
Abbreviations
AFC, 7-amino-4-trifluoromethyl coumarin; CARD, caspase activation and recruitment domain; GST, glutathione-S-transferease.
FEBS Journal 272 (2005) 1401–1414 ª2005 FEBS 1401

whilst diminished caspase-2 expression or a peptide
caspase-2 inhibitor blocked etoposide-induced cyto-
chrome crelease from mitochondria [9]. This suggests
that caspase-2 may function upstream of the mitoch-
ondrial changes associated with stress-induced apopto-
sis. This could be recapitulated in vitro [10] and has
been proposed to occur via direct caspase-2-mediated
permeabilization of mitochondrial membranes [11].
Lassus et al. found that suppression of caspase-2
expression provided equivalent protection to that con-
ferred by Apaf-1 downregulation, against apoptosis
induced by DNA damage [12]. The involvement of
caspase-2 in TRAIL-induced apoptosis has also been
reported recently, placing this enzyme upstream of Bid
cleavage in the pathway [13].
Like caspase-9, caspase-2 bears a caspase activation
and recruitment domain (CARD) in its amino-terminal
prodomain. The role of the CARD (in caspase-9 at
least) is to permit binding to aggregated adaptor pro-
teins, leading to autoactivation through ‘induced proxi-
mity’ [14]. Consistent with this, forced dimerization of
caspase-2 provoked its activation [15], and fusing the
caspase-2 prodomain to caspase-3 resulted in caspase-3
autoactivation [16]. Recent findings by Baliga et al.
indicated that dimerization is the key determinant for
initial activation of murine pro-caspase-2 [17]. The phy-
siological mechanism through which the prodomain
might trigger activation of caspase-2 is still unclear. A
molecular pathway has been proposed to link caspase-
2 to members of the tumour necrosis factor receptor
family via an adaptor molecule (RAIDD ⁄CRADD)
and intermediaries (RIP, TRADD, FADD and
TRAFs) [18,19]. However, this has not been directly
demonstrated and death ligand-mediated apoptosis
proceeds normally in caspase-2-deficient cells [7]. Other
putative caspase-2 adaptors have been proposed
[20,21], but verification of their relevance in physiologi-
cal settings has not yet been published. Tinel and
Tschopp recently reported a complex they designated
the ‘PIDD-osome’ comprising caspase-2, RAIDD and
PIDD, the formation of which promoted apoptosis
following p53-dependent DNA damage [22]. Further,
caspase-2 is recruited into a high molecular weight
complex independent of the apoptosome components
Apaf-1 and cytochrome c[23]. It has also been recently
postulated that caspase-2 may influence apoptosis [24]
and ⁄or nuclear factor-jB activation [25] through mech-
anisms unrelated to its enzymatic activity.
If caspase-2 functions as an apical caspase, it may
process and activate downstream caspases. We sought
to characterize the molecular events downstream of
human caspase-2 activation. In particular we focused
on the susceptibility of caspase-2 to suppression by
known caspase inhibitors and the ability of caspase-2
to activate effector caspases. In addition, we explored
the relationship between proteolytic processing of
caspase-2 and its enzymatic activity. Our data suggest
that processing of human caspase-2 is required for
maximal activity. Unlike other caspases, caspase-2
could not be inhibited by mammalian inhibitor of
apoptosis proteins (IAPs). Caspase-2 was able to acti-
vate caspase-7, suggesting that caspase-2 can function
as an apical caspase.
Results
High level expression of pro-caspase-2 is lethal
in yeast
Properties of caspase-2 were assessed using a yeast-
based system we have previously exploited to character-
ize other caspases and apoptotic pathways [26–28].
This system capitalizes on the observation that some
caspases kill yeast upon enforced high-level expression.
In order for caspases to kill yeast, they must both be
able to autoactivate and their proteolytic specificity
must permit cleavage of essential yeast proteins. To
assess the activity of caspase-2 in yeast, various con-
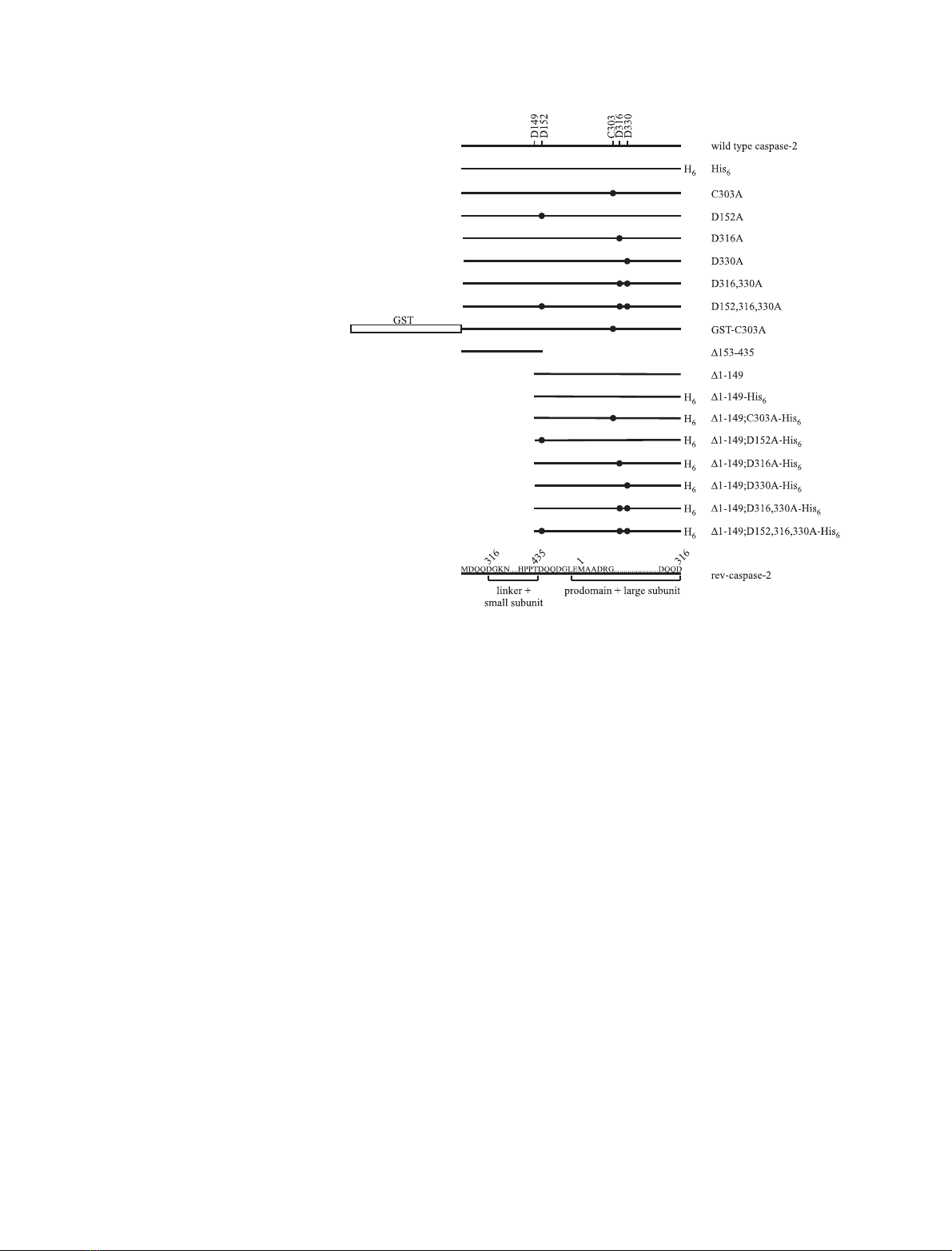
structs encoding different forms of the protein (Fig. 1)
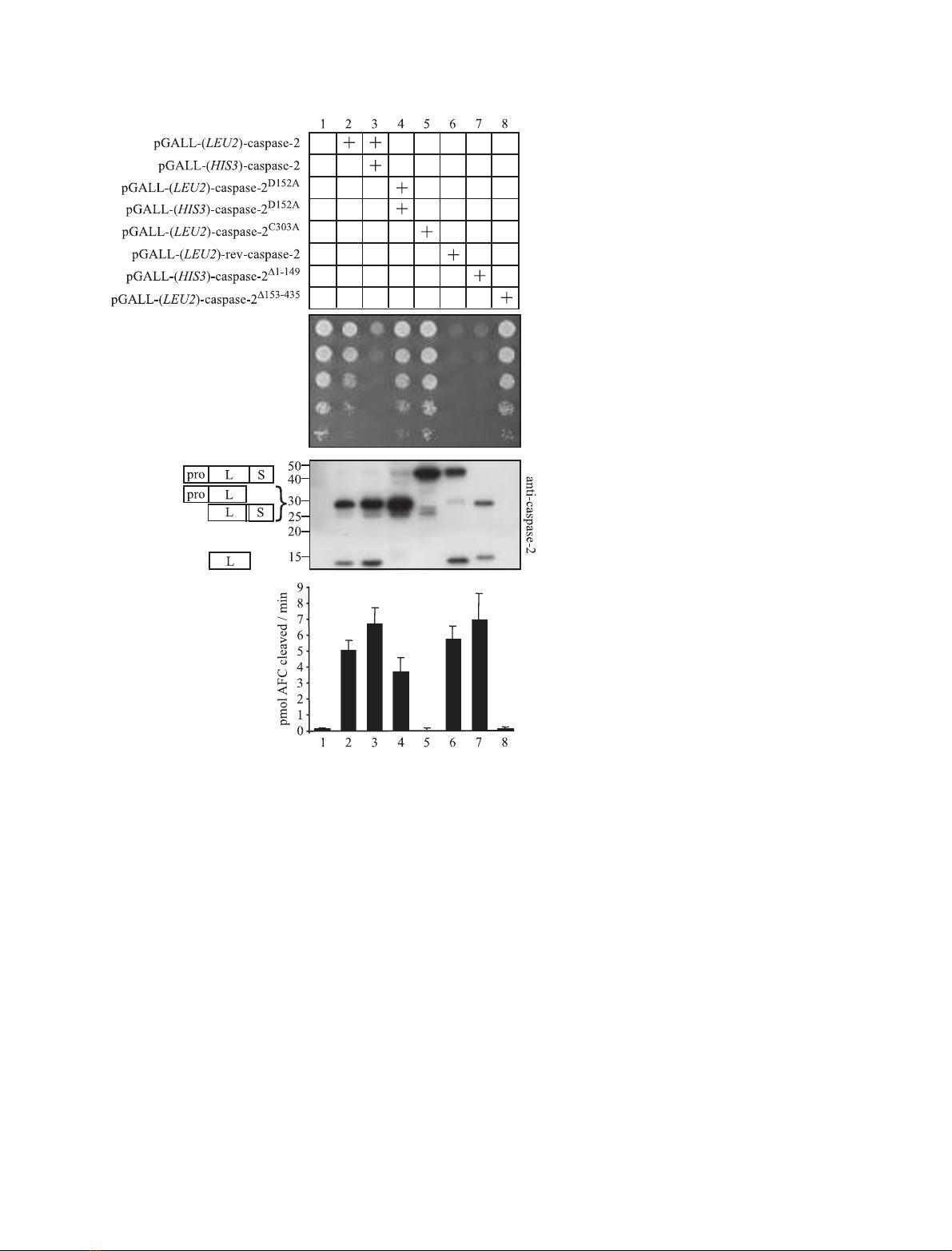
were transformed into yeast (Fig. 2A). Expression of
pro-caspase-2 using the Gal 1 ⁄10 promoter affected
yeast growth only marginally (compare growth in lane
2 to that of an empty vector transformant in lane 1).
Increasing the pro-caspase-2 expression level, by intro-
ducing an additional expression construct under dif-
ferent nutritional selection, elicited more substantial
lethality (lane 3). A caspase-2 cleavage site mutant
(D152A), from which the prodomain could not be
removed, was also expressed at a high level using two
plasmids with different nutritional selections. Com-
pared with equivalent expression of wild-type pro-
caspase-2 (lane 3), this mutant exhibited only marginal
toxicity (lane 4) suggesting that removal of the prodo-
main contributes to full enzymatic activity. Consistent
with this observation, a truncation mutant lacking
almost all of the caspase-2 prodomain (caspase-2
D1)149
)
killed yeast more efficiently than full-length caspase-2
(compare lane 7 with lane 2). An artificially autoacti-
vating version of caspase-2 (rev-caspase-2), in which
the small subunit precedes the prodomain and large
subunit [29], killed yeast readily (lane 6). The catalyti-
cally inactive mutant pro-caspase-2
C303A
was unable to
kill yeast (lane 5) implying that the lethality of wild-
type caspase-2 in yeast was due to its enzymatic activity.
The expression of the prodomain (caspase-2
D150)435
)
had no effect on yeast viability (lane 8).
Caspase-2 can activate caspase-7 and is resistant to IAPs P.-k. Ho et al.
1402 FEBS Journal 272 (2005) 1401–1414 ª2005 FEBS
To investigate the auto-processing of pro-caspase-2
in yeast, we immunoblotted lysates obtained from
yeast expressing these different forms of caspase-2 with
an antibody recognizing an epitope in the large
subunit. In lysates from yeast expressing wild-type
pro-caspase-2, a partial cleavage product was detected,
in addition to the fully processed large subunit
(Fig. 2B). Like the wild-type enzyme, the cleavage site
mutant pro-caspase-2
D152A
was processed efficiently
between the large and small subunits, however, the
mutation at D152 prevented it from being further
processed to separate the prodomain from the large
subunit. Caspase-2
C303A
remained intact as a result of
the abolished catalytic activity. Rev-caspase-2, despite
its ability to efficiently kill yeast, was only incompletely
processed. A proportion of caspase-2
D1)149
was cleaved
to remove the small subunit, thereby permitting detec-
tion of the dissociated large subunit.
The activities of these different forms of caspase-2
were also analysed biochemically using a fluorogenic
caspase-2 substrate. In this assay, the activity of an
enzyme is reflected by the efficiency with which it
cleaves the substrate to release free 7-amino-4-trifluoro-
methyl coumarin (AFC). The caspase-2-specific fluoro-
genic synthetic peptide Z-VDVAD-AFC was used as a
substrate to assess caspase-2 activity [5]. VDVADase
activity was detected in lysates from yeast expressing all
forms of caspase-2 that were capable of autoprocessing
(Fig. 2C). The most lethal forms of caspase-2 had the
highest VDVADase activity (lanes 3, 6 and 7), while ly-
sates from yeast that survived (lanes 1, 5 and 8) did not
cleave the peptide substrate. Yeast transformed with
one wild-type caspase-2 plasmid or the D152A mutant
were killed only inefficiently, however, their lysates
exhibited significant VDVADase activity. This may indi-
cate that the biochemical assay is a more sensitive meas-
ure of caspase-2 activity than the yeast death assay.
Caspase-2 is not inhibited by mammalian IAP
proteins
Members of the mammalian IAP family contribute to
the regulation of apoptotic pathways in part by their
inhibition of caspases-3, -7 and -9 [30]. Other mamma-
lian caspases (-1, -6, -8 and -10) are known to be resist-
ant to inhibition by IAPs [30], but the susceptibility of
caspase-2 to direct inhibition by IAPs has not been
reported to date. To explore the sensitivity of caspase-2
to IAP inhibition, we tested whether coexpression of
IAPs would suppress caspase-2-dependent yeast death.
Fig. 1. Schematic illustration of the caspase-
2 proteins used in this study. Mutated resi-
dues are listed above wild-type caspase-2
and are depicted with black circles.
P.-k. Ho et al. Caspase-2 can activate caspase-7 and is resistant to IAPs
FEBS Journal 272 (2005) 1401–1414 ª2005 FEBS 1403
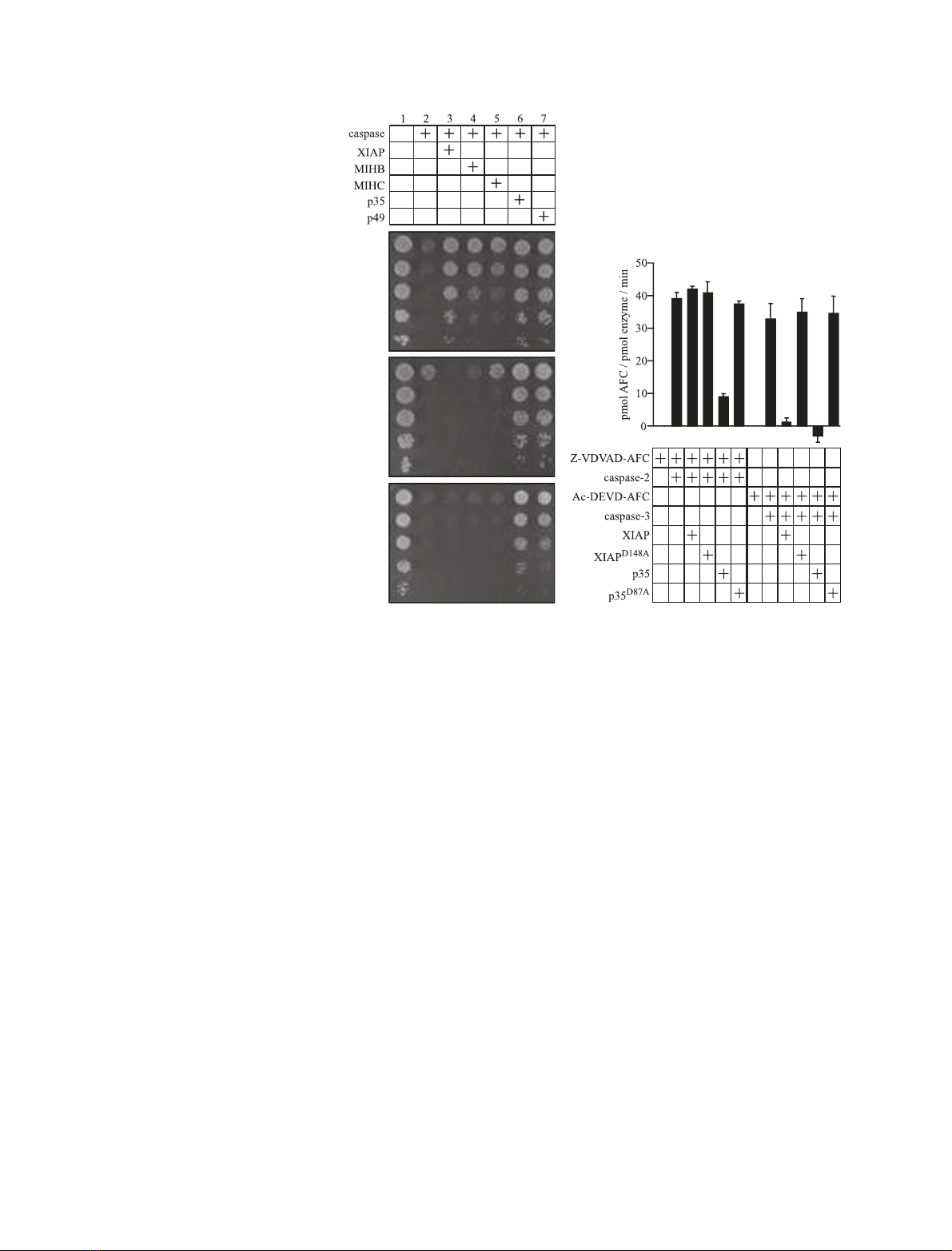
We had previously established that the inhibitors p35
and p49 could rescue yeast from caspase-2 mediated
death [31], so these baculoviral proteins were used as
positive controls. Caspase-3 effectively killed yeast and
this could be blocked by XIAP (also known as hILP),
MIHB (cIAP-1 ⁄hIAP-2 ⁄BIRC2) and MIHC (cIAP-
2⁄hIAP-1), as well as p35 and p49 (Fig. 3A). In contrast,
the mammalian IAPs could not inhibit yeast death
induced by expression either of full-length pro-caspase-2
(Fig. 3B) or of truncated caspase-2 lacking the prodo-
main (Fig. 3C). As expected, the baculoviral caspase
inhibitors p35 and p49 protected caspase-2-expressing
yeast (Fig. 3B,C).
To confirm these observations using a biochemical
approach, purified caspase-2 was mixed with recombin-
ant XIAP or the inactive mutant XIAP
D148A
[32], then
assayed for its ability to cleave the fluorogenic penta-
peptide substrate Z-VDVAD-AFC. Caspase-2 activity
was not affected by the presence of XIAP (Fig. 3D),
whereas XIAP significantly reduced the activity of
caspase-3, as demonstrated previously [33]. The pres-
ence of p35 led to a decrease in both caspase-2 and
caspase-3 activities. Inactive mutants of p35 (p35
D87A
)
and XIAP (XIAP
D148A
) were unable to inhibit either
caspase.
Caspase-2 can promote caspase-7 catalytic
activity
To explore the potential for caspase-2 to functionally
interact with other caspases, we exploited the dose-
dependent caspase-2-mediated yeast toxicity illustrated
in Fig. 2. Caspase-2 was coexpressed in yeast from a
single plasmid either alone (yielding weak lethality) or
A
B
C
Fig. 2. Caspase-2 kills yeast. (A) A semi-
quantitative assay compares the effect of
transgenes on yeast growth and viability.
Yeast cells were transformed with the indi-
cated plasmids. Suspensions of each trans-
formant were prepared at standardized
concentrations. Serial dilutions were made
and spotted onto solid inducing minimal
media vertically down the plate. Colony size
indicates growth rate and colony number
reflects cell viability. (In every experiment,
each dilution was also spotted onto a
repressing plate to verify that equivalent
numbers of each transformant were spot-
ted; data not shown). (B) Anti-caspase-2
immunoblotting of lysates from the indica-
ted transformants. The presumed identities
of each band are shown to the left (pro, pro-
domain; L, large subunit; S, small subunit).
(C) The ability of caspase-2 to cleave the flu-
orogenic peptide substrate Z-VDVAD-AFC.
Native lysates obtained from yeast were
incubated with Z-VDVAD-AFC. Fluorescence
was monitored over time and the maximal
rate of increase in free AFC was calculated
and graphed. Error bars indicate SD (n¼4).
Caspase-2 can activate caspase-7 and is resistant to IAPs P.-k. Ho et al.
1404 FEBS Journal 272 (2005) 1401–1414 ª2005 FEBS
together with the nonlethal caspases-3, -4, -6, -7 and -9
(Fig. 4A). Yeast death was used as an indicator of
caspase activity. Co-expression of caspase-2 with
caspase-7 led to a pronounced increase in yeast death,
compared to that triggered by either caspase alone
(compare lane 12 with lanes 2 and 11). Much weaker
synergy was also reproducibly observed between
caspase-2 and -3 (compare lane 6 with lanes 2 and 5).
We then tested the ability of lysates from these yeast
to cleave a fluorogenic caspase-3 substrate (Ac-DEVD-
AFC) or a caspase-2 substrate (Z-VDVAD-AFC).
Caspase-2 activity was not enhanced by coexpression
of caspases-3 or -7. However, significantly more clea-
vage of Ac-DEVD-AFC was observed when caspase-2
was coexpressed with caspase-7 (or, to a lesser extent
with caspase-3) (Fig. 4B).
To further investigate the apparent synergy between
caspase-2 and caspase-7, plasmids encoding different
forms of these enzymes were transformed into yeast in
various combinations and their effects on enzyme clea-
vage, enzyme activity and yeast growth determined
(Fig. 5). As before, high level expression of caspase-2
resulted in an active enzyme, able to efficiently kill
yeast, whereas lower expression levels of caspase-2 had
in vitro activity but weak killing activity (compare
lanes 2 and 4 in Figs 5A–C). Full length caspase-7 was
unprocessed and did not kill yeast (lane 9), whereas
caspase-7 coexpressed with caspase-2 was activated
and toxic to yeast (lane 5). The activation of caspase-7
by caspase-2 depended on caspase-2 catalytic activity
since coexpression of catalytically inactive caspase-2
with caspase-7 did not yield enzymatic activity (neither
VDVADase nor DEVDase) and did not kill yeast
(Figs 5A–C, lane 6). However, caspase-2 activation
was independent of caspase-7 as caspase-2 proteolytic
activity was the same in the presence of active or enzy-
matically inactive caspase-7 (compare Fig. 5B and C
lanes 5 and 7). Two positive controls were used for
caspase-7 activation. First, caspase-7
D1)53
, which lacks
the prodomain region and is constitutively active in
mammalian cells [34] and in yeast [35] (lane 10).
Second, as previously reported for caspase-3 [27],
caspase-7 was activated by a constitutively active
caspase-9 (rev-caspase-9) (lane 11). This autoacti-
vating caspase-9 protein, which could activate caspase-
3 [27] or caspase-7 (Fig. 5A–C, lane 11), was not able
to co-operate with caspase-2 to kill yeast (Fig. 4A, lane
14). Together, these data suggest that caspase-2 may
lie upstream of caspase-7, and not downstream of
caspase-9, in apoptotic pathways.
A
B
C
D
Fig. 3. IAPs do not inhibit caspase-2. The
caspase expression plasmids used to kill
yeast were (A) Caspase-3-lacZ (B) pGALL-
(LEU2)-caspase-2 with pGALL-(URA)-cas-
pase-2 or (C) pGALL-(URA)-caspase-2
D1)149
.
Yeast transformed with the indicated plas-
mids were spotted as described in the
legend to Fig. 1. (D) The indicated combina-
tions of caspase, fluorogenic substrate and
inhibitor were mixed together and the fluor-
escence resulting from the caspase-medi-
ated substrate cleavage was monitored and
calculated as described in the legend to
Fig. 1. Error bars indicate SD (n¼3).
P.-k. Ho et al. Caspase-2 can activate caspase-7 and is resistant to IAPs
FEBS Journal 272 (2005) 1401–1414 ª2005 FEBS 1405


























