
Entamoeba histolytica TATA-box binding protein binds
to different TATA variants in vitro
Guadalupe de Dios-Bravo
1,2
, Juan Pedro Luna-Arias
3
, Ana Marı
´a Rivero
´n
4
, Jose
´J Olivares-Trejo
5
,
Ce
´sar Lo
´pez-Camarillo
2
and Esther Orozco
5
1 Programa de Biomedicina Molecular, Escuela Nacional de Medicina y Homeopatı
´a del Instituto Polite
´cnico Nacional, Me
´xico
2 Programa de Ciencias Geno
´micas, Universidad de la Ciudad de Me
´xico, Me
´xico
3 Departamento de Biologı
´a Celular, Centro de Investigacio
´n y de Estudios Avanzados, Me
´xico
4 Departamento de Biologı
´a Molecular, Centro Nacional de Investigacio
´n Cientı
´fica (CNIC), Habana, Cuba
5 Departamento de Patologia Experimental, Centro de Investigacio
´n y de Estudios Avanzados, Me
´xico
Entamoeba histolytica is the protozoan responsible for
human amoebiasis. E. histolytica strains have distinct
capacity to damage cultured cells and human tissues
[1–4]. Expression of many molecules and cellular func-
tions involved in E. histolytica pathogenicity such as
lectins [5,6], adherence molecules [7], proteases [8,9]
and amoebapores [10] correlates with its virulence.
Variability in virulence exhibited by E. histolytica
strains might be controlled in part by transcription of
these and other virulence genes.
Transcription factors cooperate with other proteins
to regulate gene expression. First, the preinitiation
complex (PIC) is positioned around the transcription
initiation site and then, PIC interacts with other
proteins bound to upstream motifs to facilitate the
RNA polymerase II function. The absence or the pres-
ence of some nuclear factors interacting with PIC may
inhibit or promote gene expression to modulate cellu-
lar functions [11–13]. Mechanisms, molecules and
DNA sequences controlling the spatial and temporal
Keywords
Entamoeba histolytica;K
D
; promiscuous
DNA-binding activity; TATA-binding protein;
TATA variants
Correspondence
Esther Orozco, Departamento de Patologı
´a
Experimental, Centro de Investigacio
´nyde
Estudios Avanzados, IPN. C. P. 07360,
Me
´xico, D. F.
Fax: +52 55 57477108
Tel: +52 55 50613800 ext 5642
E-mail: esther@mail.cinvestav.mx
(Received 23 June 2004, revised 8 December
2004, accepted 11 January 2005)
doi:10.1111/j.1742-4658.2005.04566.x
The ability of Entamoeba histolytica TATA binding protein (EhTBP) to
interact with different TATA boxes in gene promoters may be one of the
key factors to perform an efficient transcription in this human parasite. In
this paper we used several TATA variants to study the in vitro EhTBP
DNA-binding activity and to determine the TATA-EhTBP dissociation
constants. The presence of EhTBP in complexes formed by nuclear extracts
(NE) and the TATTTAAA oligonucleotide, which corresponds to the
canonical TATA box for E. histolytica, was demonstrated by gel-shift
assays. In these experiments a single NE-TATTTAAA oligonucleotide
complex was detected. Complex was retarded by anti-EhTBP Igs in super-
shift experiments and antibodies also recognized the cross-linked complex
in Western blot assays. Recombinant EhTBP formed specific complexes
with TATA variants found in E. histolytica gene promoters and other
TATA variants generated by mutation of TATTTAAA sequence. The dis-
sociation constants of recombinant EhTBP for TATA variants ranged
between 1.04 (±0.39) ·10
)11
and 1.60 (±0.37) ·10
)10
m. TATTTAAA
and TAT_ _AAA motifs presented the lowest K
D
values. Intriguingly, the
recombinant EhTBP affinity for TATA variants is stronger than other
TBPs reported. In addition, EhTBP is more promiscuous than human and
yeast TBPs, probably due to modifications in amino acids involved in
TBP-DNA binding.
Abbreviations
EhTBP, Entamoeba histolytica TATA-box binding protein; EMSA, electrophoretic mobility shift assays; rEhTBP, recombinant Entamoeba
histolytica TATA-box binding protein; NE, nuclear extracts.
1354 FEBS Journal 272 (2005) 1354–1366 ª2005 FEBS

transcription patterns during growth, differentiation
and development have been widely studied [14,15].
In eukaryotes, general transcription factors such as
TFIID ⁄TFIIB, TFIIA, TFIIE, TFIIF ⁄RNA poly-
merase II and TFIIH are assembled on the core pro-
moter before transcription begins [16,17].
The TATA binding protein (TBP) is the first fac-
tor that binds DNA to recruit proteins on PIC and
initiate gene transcription [18,19]. Mammalian TBPs
can productively bind to a large number of diverse
TATA elements. An exhaustive statistical genomic
survey documented that the TATA box is an A ⁄T-
rich 8 bp segment, often flanked by G ⁄C-rich
sequences [20].
Certain E. histolytica genes are activated or down
regulated during liver abscesses production by tro-
phozoites [9] and during epithelia colonization and
invasion. However, we ignore which transcription
factors modulate these events and others related to
the parasite survival such as trophozoites differenti-
ation into cysts. Few transcription factors have been
detected and cloned in E. histolytica. URE3-BP,
EhEBP1 and EhEBP2 proteins regulate the hgl5 gene
expression [21,22] and an EhC ⁄EBP-like protein is
involved in EhPgp1 gene activation [23,24]. Addition-
ally, Ehtbp [25] and Ehp53 [26] have been character-
ized as the orthologous of the mammalian tbp and
p53 genes, respectively. The E. histolytica TATA-
binding protein (EhTBP) is the only member of the
basal transcription machinery cloned and character-
ized in this parasite.
The EhTBP functional DNA-binding domain has
55% homology with human TBP, whereas the
EhTBP N-terminal domain is rich in hydrophobic
residues and quite different from mammalian TBPs
[25]. EhTBP C-terminus displays a predicted protein
structure fitting to the crystallized human TBP
[27,28], but its biological activity has been poorly
studied.
E. histolytica gene promoters have the TATTTAAA
sequence, which is considered as the canonical TATA
box for EhTBP [29]. However, EhTBP binding to this
sequence has not been fully demonstrated. In addition,
TATTTAAA variants have been found surrounding
the core promoter, suggesting that these motifs could
also act as TATA boxes [30,31], but EhTBP affinity
for these sequences is also unknown. In this paper, we
studied the in vitro EhTBP binding affinity for differ-
ent TATA sequences found in E. histolytica gene pro-
moters and others designed by us producing mutations
in the TATTTAAA sequence. We also calculated the
K
D
of recombinant E. histolytica TBP (rEhTBP) for
several TATA variants.
Results
E. histolytica nuclear extracts (NE) and TATTT-
AAA(1) oligonucleotide form specific complexes
Bruchhaus et al. [29] using NE in EMSA and doing
in silico analysis proposed that the TATTTAAA
sequence is the consensus TATA box for E. histolytica.
On the other hand, Luna-Arias et al. [25] showed the
homology of EhTBP with human TBP. However,
the presence of EhTBP in complexes formed with
E. histolytica NE and TATTTAAA(1) oligonucleotide
has not been directly demonstrated yet.We first investi-
gated the presence of EhTBP in the complex formed by
E. histolytica NE and TATTTAAA(1) oligonucleotide
by supershift, cross-linking and Western blot assays.
When incubated with fresh NE, TATTTAAA(1) oligo-
nucleotide migration was retarded, forming a single
band (Fig. 1A, lane 1). The NE-TATTTAAA(1) com-
plex was specifically competed by TATTTAAA(1) cold
oligonucleotide (Fig. 1A, lane 2), whereas it remained
when double-stranded poly(dG-dC) or TtTTTttt(7)
oligonucleotide were used as unspecific competitors
(Fig. 1A, lanes 3 and 4, respectively). The presence of
EhTBP in this complex was evidenced in supershift
assays by anti-rEhTBP Igs. Two bands appeared when
1lL of antibodies was added to the mixture (Fig. 1B,
lane 3). The lower band comigrated with that formed
by NE and TATTTAAA(1) oligonucleotide, whereas
the other band migrated slower, due to the partial
supershift produced by the antibody. When 5 lL
of anti-rEhTBP Igs were added to the mixture, the
complex was completely disrupted (Fig. 1B, lane 4), as
it has been reported for other supershift experiments
[32]. Anti-E. histolytica actin antibodies had no effect
on the complex formed (Fig. 1C, lane 2).
In cross-linking assays, using a UV-irradiated mix-
ture of E. histolytica NE and TATTTAAA(1) oligonu-
cleotide, we distinguished a radioactive DNA–protein
band of 50 kDa (Fig. 1D, lane 5). This band may be
formed by the radioactive probe (11 kDa) bound to
endogenous EhTBP (26 kDa) and other protein cross-
linked to the complex. As expected, the 50 kDa radio-
active band was competed by TATTTAAA(1) cold
oligonucleotide (Fig. 1D, lane 6), but it remained in
the presence of the poly (dG-dC) unspecific competitor
(Fig. 1D lane 7). No complexes were detected in lanes
with either nonirradiated or irradiated free probe, or
with the nonirradiated oligonucleotide-NE mixture
(Fig. 1D, lanes 2–4, respectively). In Western blot
assays of UV cross-linked DNA–protein complexes,
anti-rEhTBP Igs recognized the radioactive 50 kDa
band. This confirms that EhTBP is part of the complex
G. de Dios-Bravo et al. Promiscuous E. histolytica TBP
FEBS Journal 272 (2005) 1354–1366 ª2005 FEBS 1355
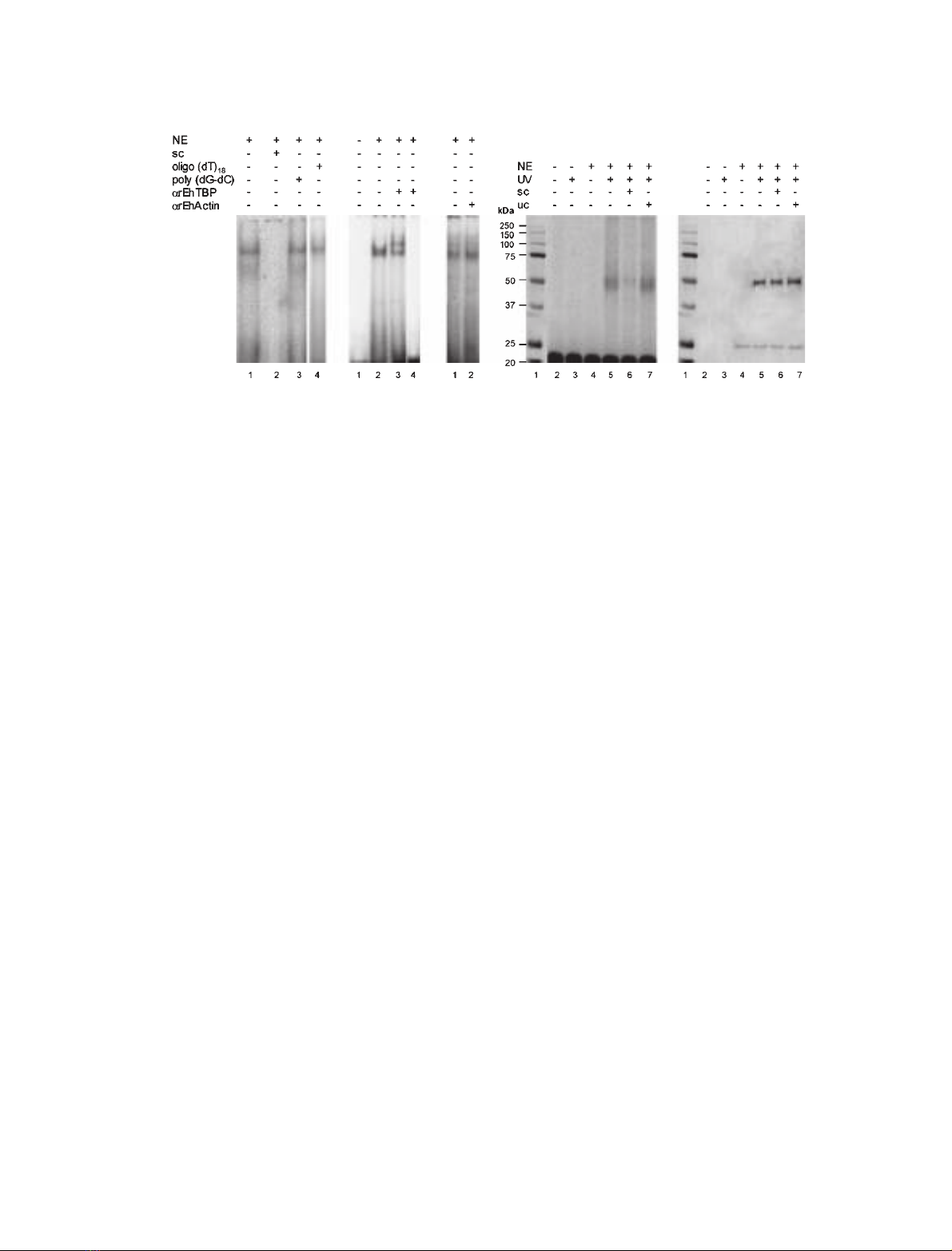
probably associated to another 13 kDa unknown
protein (Fig. 1E, lanes 5–7). The antibodies also recog-
nized the same band in the lane where TATTTAAA(1)
cold oligonucleotide was used as specific competitor
(Fig. 1E, lane 6). As expected, in this lane the complex
was formed by the irradiated cold oligonucleotide and
NE mixture. Unbound 26 kDa EhTBP comigrated in
the gel with the 25 kDa marker (Fig. 1E, lanes 4–7).
Data from these experiments altogether demonstrated
the presence of EhTBP in NE-TATTTAAA(1) oligo-
nucleotide complexes.
Recombinant EhTBP binds to TATTTAAA(1)
oligonucleotide
The rEhTBP was expressed in bacteria as a His
6
-
tagged 30 kDa polypeptide. rEhTBP was purified by
affinity chromatography and its integrity and identity
were verified by Coomassie blue stained gels
(SDS ⁄PAGE) (Fig. 2A) and Western blot assays using
anti-rEhTBP Igs (Fig. 2B). In EMSA, purified rEhTBP
formed a single band with TATTTAAA(1) probe
(Fig. 2C,lane 2).The complex was competed by cold
TATTTAAA(1) oligonucleotide, whereas it remained
in the presence of poly (dG-dC) unspecific competitor
(Fig. 2C, lanes 3 and 4). To discard endogenous
TATA binding activity in bacterial extracts, we tested
by EMSA the capacity of induced and noninduced
bacterial extracts to form complexes with TATTT
AAA(1) probe. Results showed that complex was only
formed with extracts of induced bacteria expressing
rEhTBP (Fig. 2D,lane 3) whereas noninduced bacteria
did not form any complexes with TATTTAAA(1)
oligonucleotide (Fig. 2D, lane 2).
DNA-binding activity of purified rEhTBP
for TATA variants
In humans and other organisms, variants of the canon-
ical TATA box have been reported to be functional
[33,34]. On the other hand, the TATTTAAA sequence
and several variants are found in many E. histolytica
gene promoters at )20 to )40 bp upstream the tran-
scription initiation site [30], although other TATA
variants have been experimentally found at longer
distances in Ehtbp and EhRabB genes (our unpublished
data). We studied the binding activity of rEhTBP for
different TATA sequences present in gene promoters
(oligonucleotides TATTTAAA(1), TAT_ _AAA(4),
TAT_ _AAg(5) and TATTaAAA(6)), and for mutated
versions of TATTTAAA(1) probe [oligonucleotides
TAgTgAAA(2) and TATTggAA(3)] (Table 1). We
ABCD E
Fig. 1. Binding of nuclear extracts to TATTTAAA(1) oligonucleotide. (A) NE (25 lg) and [
32
P]ATP[cP] end-labeled TATTTAAA(1) oligonucleotide
(10 000 c.p.m., 157 pM) were incubated for 15 min at 4 C for EMSA as described in Experimental procedures. Lane 1, no competitor; lane
2, 300-fold molar excess of unlabeled TATTTAAA(1) oligonucleotide as specific competitor (sc); as unspecific competitors we added 300-fold
molar excess of: lane 3, poly(dG-dC) and, lane 4, oligo(dT)
18
. (B) Supershift gel assay using purified anti-rEhTBP Igs. EMSA were performed
as above, except that before adding the labeled oligonucleotide, the mixture was preincubated with: lane 1, no NE; lane 2, no antibody; lane
3, 1 lL of purified anti-rEhTBP Igs; lane 4, 5 lL of anti-rEhTBP Igs. (C) Supershift gel assay performed as in B, but using anti-E. histolytica
actin Igs. Lane 1, no antibody; lane 2, 5 lL of anti-E. histolytica actin Ig. (D) UV-cross-linking assay of NE (60 lg) and TATTTAAA(1) (50 000
c.p.m., 785 pM). Mixtures for EMSA were UV irradiated at 320 nm for 10 min at 4 C, analyzed by 12% SDS ⁄PAGE and radioactivity was
determined as described in Experimental procedures. Lane 1, molecular mass markers; lane 2, nonirradiated free probe; lane 3, irradiated
free probe; lane 4, nonirradiated NE-oligonucleotide mixture; lane 5, irradiated NE-oligonucleotide mixture; lane 6, irradiated NE-oligonucleo-
tide mixture containing 300-fold molar excess of unlabeled TATTTAAA(1) oligonucleotide as specific competitor (sc); lane 7, irradiated
NE-oligonucleotide mixture containing 300-fold molar excess of poly (dG-dC) as unspecific competitor (uc). (E) Western blot assay of UV
cross-linked DNA–protein complexes shown in D, using anti-rEhTBP Igs.
Promiscuous E. histolytica TBP G. de Dios-Bravo et al.
1356 FEBS Journal 272 (2005) 1354–1366 ª2005 FEBS
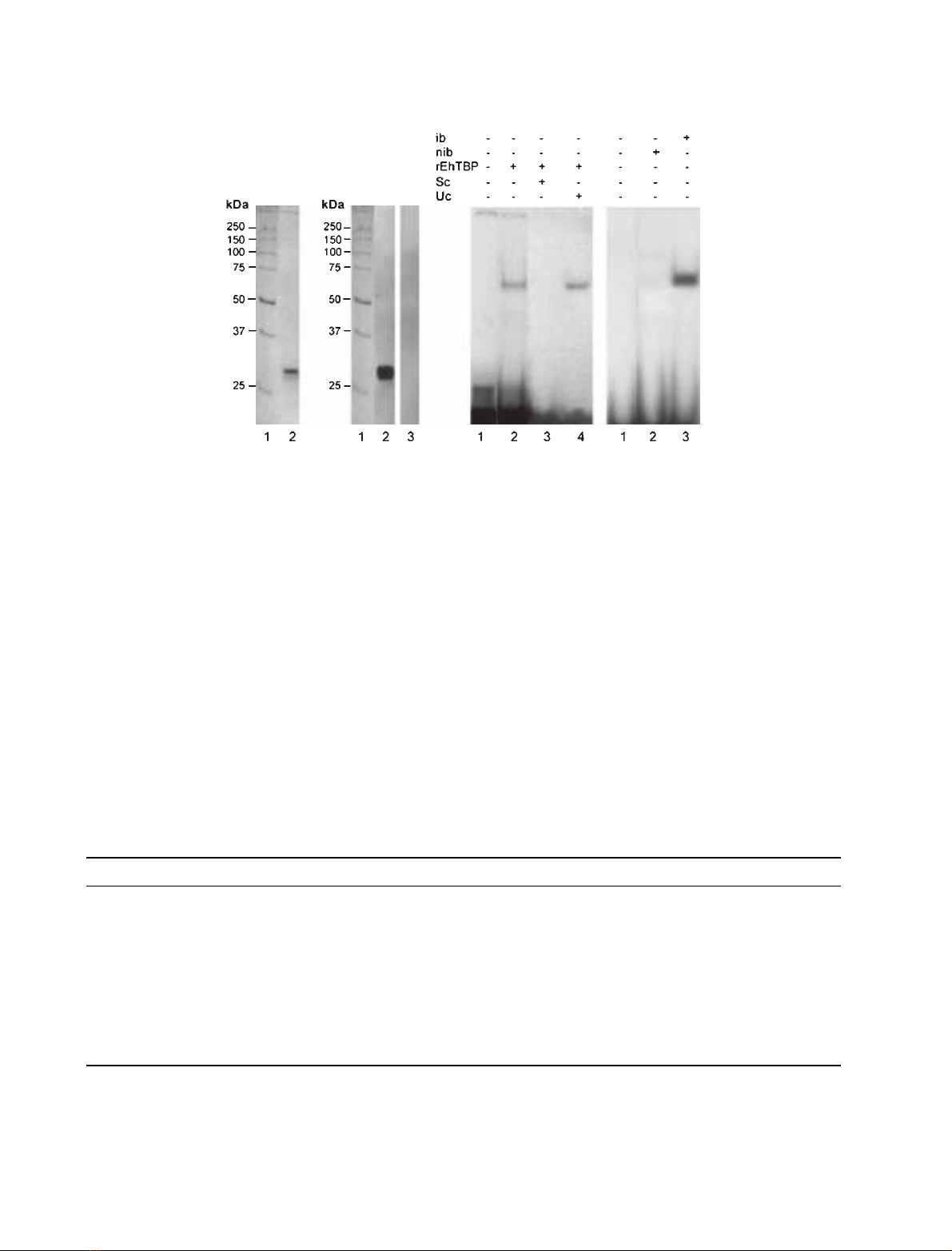
introduced g’s in the third, fifth and sixth positions of
the TATTTAAA(1) sequence, because these positions
have been reported as important for DNA-binding
activity for human and yeast TBPs [33,34].
DNA-binding activity of rEhTBP for distinct TATA
oligonucleotides was evaluated by EMSA using
433 nm(over-saturating concentration) of purified
rEhTBP and 10 000 c.p.m. (157 pm) of the probes.
Figure 3 displays experiments showing that rEhTBP
specifically binds to all oligonucleotides tested. Com-
plexes formed by rEhTBP and TATA box variants
were fully competed by the same probe and by TAT-
TTAAA(1) oligonucleotide (Fig. 3). In these assays,
two complexes were observed with TAT_ _AAA(4)
and TAT_ _AAg(5) probes, which were specifically
competed by TATTTAAA(1) oligonucleotide and by
the same probe. The presence of two complexes in
some experiments could be due to conformational
Table 1. Positions of TATTTAAA (1) sequence and putative TATA variants in E. histolytica gene promoters.
TATA variants Gene promoter First nucleotide location
b
Reference
(12 3456 78)
a
5’-T A T T T A A A-3’ (1) EhPgp5
Ehactin
(-31)
c
(-30)
d
[47]
(http://www.sanger.ac.uk/Projects/E_histolytica)
5’-T A gTgA A A-3’ (2) Not found
5’-T A T T ggA A-3’ (3) Not found
5’-T A T __A A A-3’ (4) Ehtbp (-109)
c
(Unpublished)
Ehtub1 (-27)
d
(http://www.sanger.ac.uk/Projects/E_histolytica)
EhRabB (-44)
c
(Unpublished)
5’-T A T __AAg-3’ (5) Ehenol (-50)
d
(http://www.sanger.ac.uk/Projects/E_histolytica)
5’-T A T T a A A A-3’ (6) Ehpfo (-31)
c
[48]
a
Numbers show the base composition in TATA variants.
b
Nucleotide position is referred to the experimentally
c
and in silico
d
determined
transcription initiation sites. Putative TATA boxes are defined as TATA sequences upstream of the ATTCA ⁄G, ATCA or ACGC consensus
transcription initiation sites.
AB C D
Fig. 2. Immunodetection of rEhTBP, and EMSA of TATTTAAA(1) and rEhTBP. rEhTBP was produced by IPTG induced bacteria transformed
with the full length Ehtbp gene cloned in pRSET A and purified through nickel NTA-agarose columns as described in Experimental proce-
dures. (A) Coomassie blue stained gel (12% SDS ⁄PAGE) of purified rEhTBP under native conditions. Lane 1, molecular mass markers; lane
2, purified rEhTBP. (B) Western blot assay of purified rEhTBP using anti-rEhTBP Igs. Lane 1, molecular weight markers; lane 2, stripe
sequentially incubated with anti-rEhTBP Igs and peroxidase-coupled goat anti-rabbit secondary Igs; lane 3, as in lane 2 but anti-rEhTBP Igs
were omitted. (C) EMSA of purified rEhTBP with TATTTAAA(1) oligonucleotide as described in Experimental procedures. Lane 1, free probe;
lane 2, no competitor; lane 3, 300-fold molar excess of unlabeled TATTTAAA(1) probe as specific competitor (sc); lane 4, 300-fold molar
excess of unspecific competitor (uc). (D) EMSA using 15 lg of bacterial extracts. Lane 1, free probe; lane 2, non induced bacteria (nib) carry-
ing pRSET A-Ehtbp plasmid; lane 3, induced bacteria (ib) expressing rEhTBP.
G. de Dios-Bravo et al. Promiscuous E. histolytica TBP
FEBS Journal 272 (2005) 1354–1366 ª2005 FEBS 1357
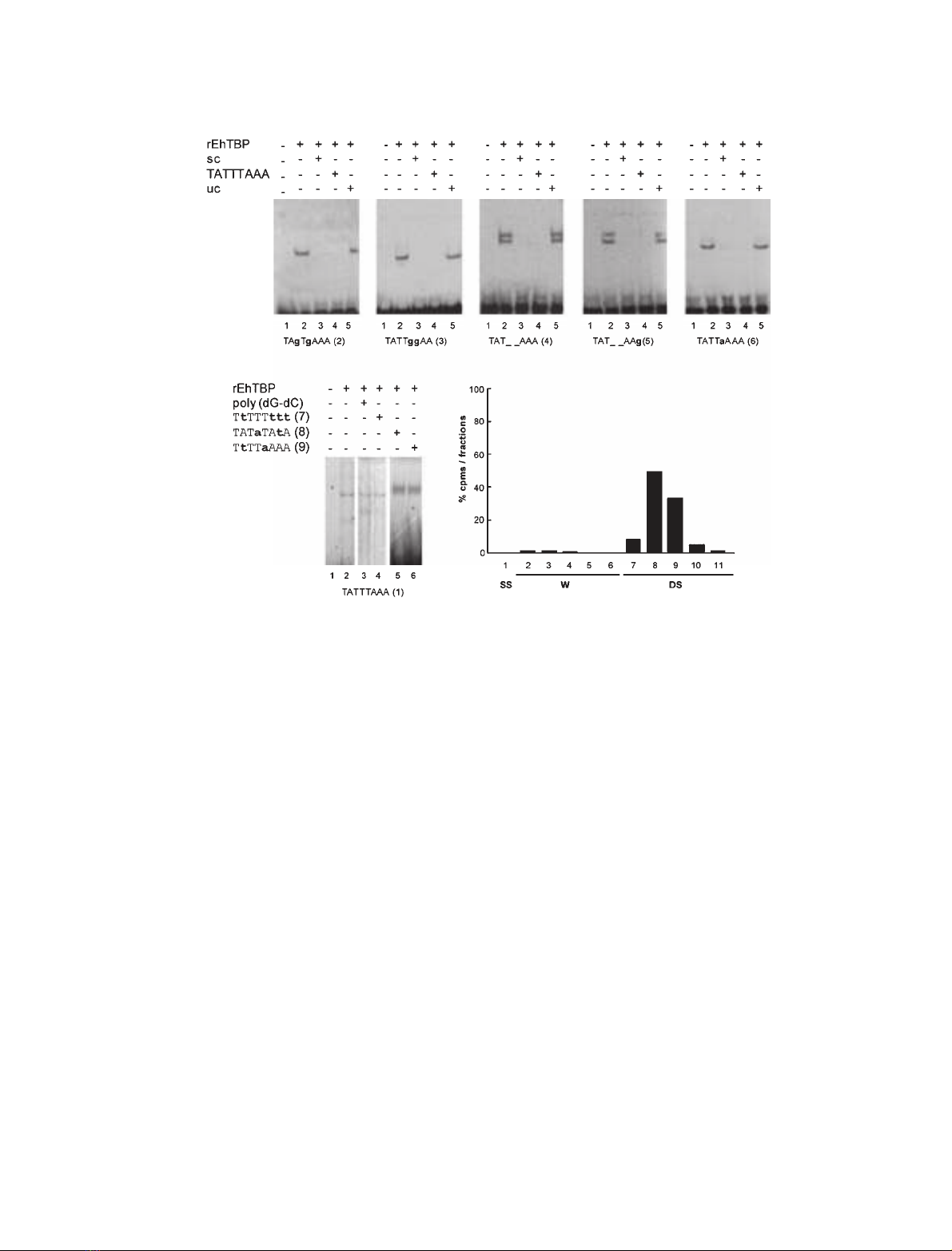
changes of the DNA–protein complex, which may
affect its electrophoretic migration. To discard the pos-
sibility that rEhTBP could bind to any AT rich
sequence, we performed a shift assay with rEhTBP
and [
32
P]ATP[cP] end-labeled double stranded
TtTTTttt(7), TATaTAtA(8) or TtTTaAAA(9) oligonu-
cleotides. rEhTBP did not bind to these sequences
(data not shown), indicating that rEhTBP is not
merely an AT-rich DNA binding protein without dis-
crimination capacity. Obviously, these oligonucleotides
did not compete the complex formed with TATTT-
AAA(1) and rEhTBP (Fig. 3F). We also verified that
in our experiments, rEhTBP was indeed bound to dou-
ble-stranded oligonucleotides and not to free labeled
single-stranded probes. Labeled probes were passed
through a hydroxyapatite column and c.p.m. were
counted in the unbound and eluted fractions. In all
cases, more than 99% of the radioactivity was found
bound to the hydroxyapatite column and it was eluted
with 0.4 mphosphate buffer. Figure 3G shows the elu-
tion profile for TATTTAAA(1) oligonucleotide as a
representative experiment. All together these results
showed that rEhTBP has an in vitro binding capacity
for distinct TATA elements.
Quantification of rEhTBP DNA-binding activity
for different TATA oligonucleotides
Binding activity of rEhTBP for TATA variants was
quantified (as described in Experimental procedures) at
A
FG
BCD
E
Fig. 3. rEhTBP specifically binds to TATTTAAA(1) oligonucleotide and TATA variants. (A–E) Purified rEhTBP (433 nM) was incubated with
[
32
P]ATP[cP] end-labeled TATA variants (10 000 c.p.m., 157 pM) for EMSA as described in Experimental procedures. Lane 1, free probe; lane
2, no competitor; lane 3, competition with 300-fold molar excess of the same TATA variant as specific competitor (sc); lane 4, competition
with 300-fold molar excess of TATTTAAA(1) oligonucleotide; lane 5, competition with 300-fold molar excess of unspecific competitor (uc).
The TATA oligonucleotide used in each case is shown below each gel. (F) Control binding assay of rEhTBP (433 nM) with 157 pM(10 000
c.p.m.) of TATTTAAA(1) probe. Lane 1, TATTTAAA(1) free probe; lane 2, purified rEhTBP incubated with TATTTAAA(1) probe; lane 3, purified
rEhTBP preincubated with 300-fold molar excess of unlabeled poly (dG-dC) before adding the labeled TATTTAAA(1) probe; lane 4, unlabeled
TTTTTTTT(7) oligonucleotide; lane 5, unlabeled TATATATA(8) oligonucleotide, and lane 6 TTTTAAAA(9) oligonucleotide were used as unspe-
cific competitors. (G) Elution profile of labeled TATTTAAA (1) probe passed through a hydroxyapatite column as described in Experimental
procedures. Fraction 1, unbound single stranded DNA (SS); fractions 2–6, washes with 2.5 mL of 0.12 Mphosphate buffer pH 6.8 (W); frac-
tions 7–11, elution with 2.5 mL of 0.4 Mphosphate buffer pH 6.8 (DS). Volume of each fraction was 0.5 mL. Radioactivity was represented
as percentage of the total radioactivity (30 000 c.p.m.) loaded into the column.
Promiscuous E. histolytica TBP G. de Dios-Bravo et al.
1358 FEBS Journal 272 (2005) 1354–1366 ª2005 FEBS


























