
CpG methylation of the CENP-B box reduces human
CENP-B binding
Yoshinori Tanaka
1,2,
*, Hitoshi Kurumizaka
1,3
and Shigeyuki Yokoyama
1,2,4
1 Protein Research Group, RIKEN Genomic Sciences Center, Suehiro-cho, Tsurumi, Yokohama, Japan
2 Department of Biophysics and Biochemistry, Graduate School of Science, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo, Japan
3 Waseda University School of Science and Engineering, 3-4-1 Okubo, Shinjuku-ku, Tokyo, Japan
4 RIKEN Harima Institute at Spring-8, 1-1-1 Kohto, Mikazuki-cho, Sayo, Hyogo, Japan
The centromere of eukaryotic chromosomes plays an
essential role in the proper segregation of chromo-
somes at mitosis and meiosis, and has a special hetero-
chromatin structure, which is composed of a-satellite
DNA repeats and their associated proteins. The
human centromere proteins A, B and C (CENP-A,
CENP-B and CENP-C, respectively) are such centro-
mere-specific DNA-binding proteins [1–7]. Neither
CENP-A nor CENP-C shows any sequence specificity
in DNA binding. In contrast, CENP-B is known to
specifically bind a 17 base-pair sequence (the CENP-B
box), which appears in every other a-satellite repeat
(171 base-pairs) in human centromeres [8–10].
CENP-B is an 80 kDa protein that contains DNA-
binding and dimerization domains at the N-terminus
and C-terminus, respectively [11–13]. Biochemical ana-
lyses with nucleosomes reconstituted in vitro suggested
that the CENP-B box sequence functions as a cis
element for centromere-specific nucleosome assembly
[14]. In vivo analyses with cultured human cells
Keywords
CENP-B; centromere; DNA methylation;
chromatin; heterochromatin
Correspondence
2
H. Kurumizaka, Waseda University School of
Science and Engineering, 3-4-1 Okubo,
Shinjuku-ku, Tokyo 169-8555, Japan
Fax: +81 3 5292 9211
Tel: +81 3 5286 8189
E-mail: kurumizaka@waseda.jp
S. Yokoyama, Protein Research Group,
RIKEN Genomic Sciences Center, 1-7-22
Suehiro-cho, Tsurumi, Yokohama 230-0045,
Japan
Fax: +81 45 503 9195
Tel: +81 45 503 9196
E-mail: yokoyama@biochem.s.u-tokyo.ac.jp
*Present address
Toray Industries, Inc. New Frontiers
Research Laboratories, 1111 Tebiro, Kamak-
ura, Kanagawa 248–0036, Japan
(Received 15 September 2004, accepted
1 October 2004)
doi:10.1111/j.1432-1033.2004.04406.x
In eukaryotes, CpG methylation is an epigenetic DNA modification that is
important for heterochromatin formation. Centromere protein B (CENP-
B) specifically binds to the centromeric 17 base-pair CENP-B box DNA,
which contains two CpG dinucleotides. In this study, we tested complex
formation by the DNA-binding domain of CENP-B with methylated and
unmethylated CENP-B box DNAs, and found that CENP-B preferentially
binds to the unmethylated CENP-B box DNA. Competition analyses
revealed that the affinity of CENP-B for the CENP-B box DNA is reduced
nearly to the level of nonspecific DNA binding by CpG methylation.
Abbreviations
CENP-B, centromere protein B; RNAi, RNA interference; siRNA, small interfering RNA.
282 FEBS Journal 272 (2005) 282–289 ª2004 FEBS
revealed that the presence of the CENP-B box is essen-
tial for the formation of functional minichromosomes
[15,16]. Therefore, the CENP-B box is required for the
formation of a functional centromere. However,
CENP-B null mice appeared to be normal [17–19].
This discrepancy about the CENP-B dispensibility for
mouse development and the CENP-B box requirement
for minichromosome maintenance in human cells may
be explained by presuming the existence of functional
homologues of CENP-B. In fact, CENP-B-like pro-
teins have been identified in humans, and the func-
tional redundancy of CENP-B homologues has also
been found in the fission yeast Schizosaccharomyces
pombe [20–22].
In eukaryotic cells, methylation of the cytosine
within the CpG dinucleotide is an epigenetic DNA
modification that is important for heterochromatin
formation. The human a-satellite consensus sequence
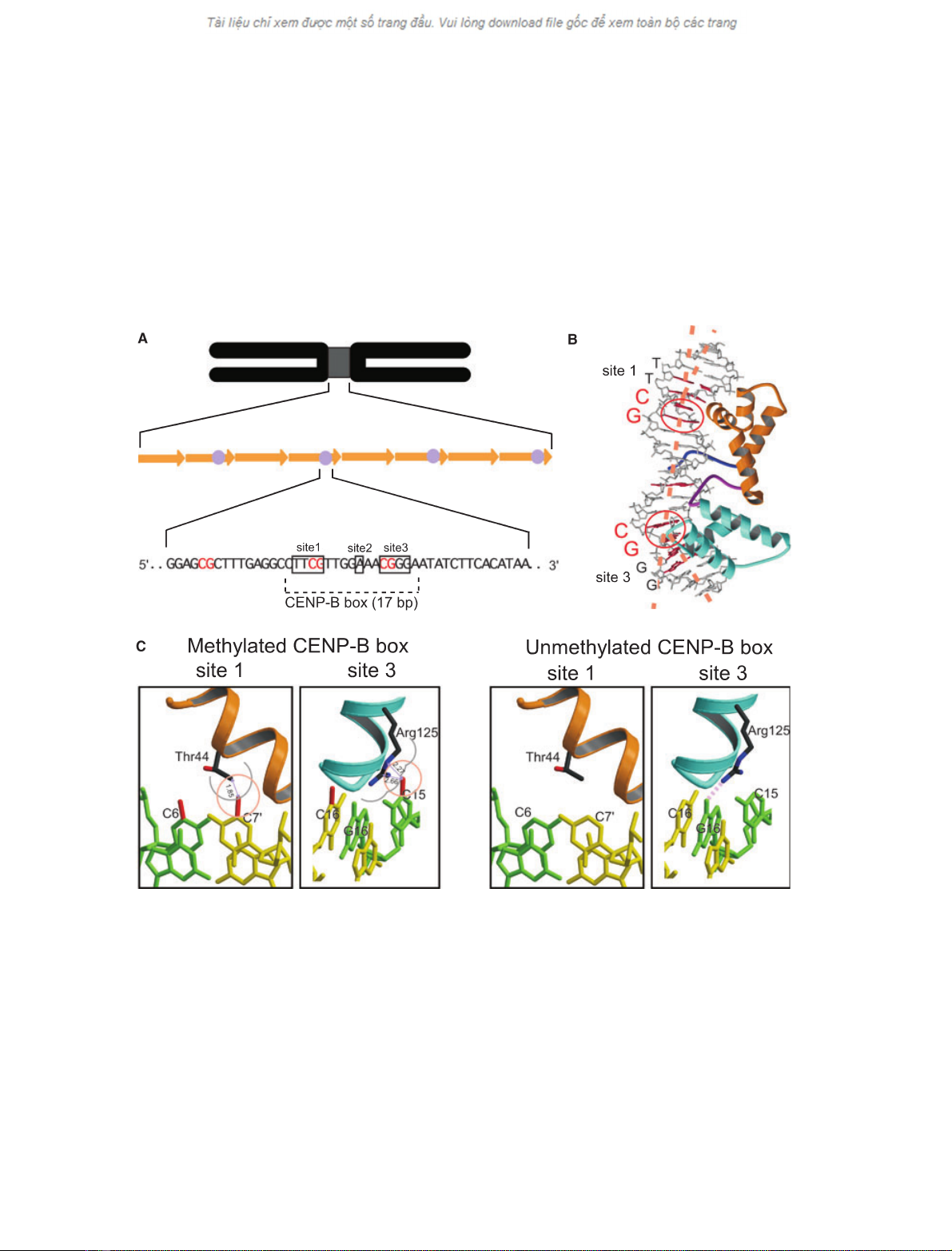
Fig. 1. Human a-satellite DNA and the CENP-B(1–129)–DNA complex structure. (A) Schematic diagram showing the organization of human
a-satellite DNA and the CENP-B box. Arrows and circles in the middle row indicate a-satellite DNA repeats and CENP-B boxes, respectively.
The a-satellite consensus sequence [23], containing a 17 bp CENP-B box, is shown in the bottom row. The three boxes, marked as sites 1,
2 and 3, indicate the essential bases for CENP-B binding to the CENP-B box DNA. The three CpG sequences in the a-satellite sequence are
shown in red. (B) The CENP-B(1–129)–DNA complex structure [25]. The essential bases for CENP-B binding are coloured red in the model
structure. The CpG sites that are sharply kinked by the CENP-B binding are encircled in red. The nucleotide sequences of sites 1 and 3 are
indicated in the left column. (C) Model for the interaction between CENP-B and the CpG-methylated CENP-B box. (Left) Interaction between
CENP-B and the methylated CENP-B box DNA at sites 1 and 3. The methyl groups, which are modelled on the C5 atoms of cytosines in the
CpG dinucleotides, are coloured red. The van der Waals radii of the methyl groups (2.0 A
˚) are shown in orange circles. The van der Waals
radii of the Thr44 and Arg125 side chains are shown in grey circles. Distances between the cytosine methyl groups and the nearest Thr44
and Arg125 side chain atoms are indicated by arrows. (Right) Interaction between CENP-B and the unmethylated CENP-B box DNA at sites
1 and 3. The pink dotted line in the right panel indicates the hydrogen bond between the side chain NH
2
group of Arg125 and the N7 atom
of guanine.
Y. Tanaka et al.CpG methylation reduces human CENP-B binding
1
FEBS Journal 272 (2005) 282–289 ª2004 FEBS 283
contains only three CpG sequences within its 171 base-
pair sequence [23]. Interestingly, two of the three CpG
sequences in the a-satellite consensus sequence
are located within site 1 (5¢-pTpTpCpG-3¢) and site 3
(5¢-pCpGpGpG-3¢) of the CENP-B box (Fig. 1A; [9]).
Demethylation of satellite DNA sequences, accom-
plished by growing cells in the presence of the DNA
methyltransferase inhibitor, 5-aza-2¢-deoxycytidine,
resulted in the redistribution of CENP-B [24], indica-
ting that the CpG methylation affects the CENP-
B–DNA interaction. Our structural analysis of the
CENP-B DNA-binding domain [CENP-B(1–129)]
complexed with the CENP-B box DNA revealed that
CENP-B induced sharp kinks at the CpG sequences of
sites 1 and 3 upon DNA binding (Fig. 1B; [25]). There-
fore, CpG methylation may regulate CENP-B binding
to the CENP-B box DNA within centromeric a-satel-
lite repeats.
In this study, we tested the complex formation of
CENP-B(1-129) with methylated and unmethylated
CENP-B box DNAs by a complex-reconstitution
assay, and found that the affinity of CENP-B for CpG
methylated CENP-B box DNA is significantly reduced.
Results
Model for the interaction between CENP-B and
the CpG-methylated CENP-B box
In eukaryotes, DNA methylation occurs at the C5
atom of cytosine by the action of methyltransferases.
To evaluate the effect of CpG methylation on the
CENP-B–DNA interaction, we modeled additional
methyl groups at the C5 atoms of site 1 (cytosine 6 and
cytosine 7¢) and site 3 (cytosine 15 and cytosine 16¢)in
the CENP-B(1–129)–DNA complex structure (Fig. 1C).
In the crystal structure of the CENP-B(1–129)–DNA
complex [25], the CpG sequences in sites 1 and 3 of the
CENP-B box DNA were sharply kinked by CENP-
B(1–129) binding (16for site 1 and 43for site 3;
Fig. 1B). As shown in Fig. 1C, additional methyl
groups on cytosine 7¢and cytosine 15 caused steric cla-
shes with the side chains of Thr44 and Arg125, respect-
ively. Therefore, the CENP-B(1–129)–methylated DNA
complex model suggested that the methylations of cyto-
sine 7¢and cytosine 15 may sterically interfere with
CENP-B binding to the CENP-B box DNA.
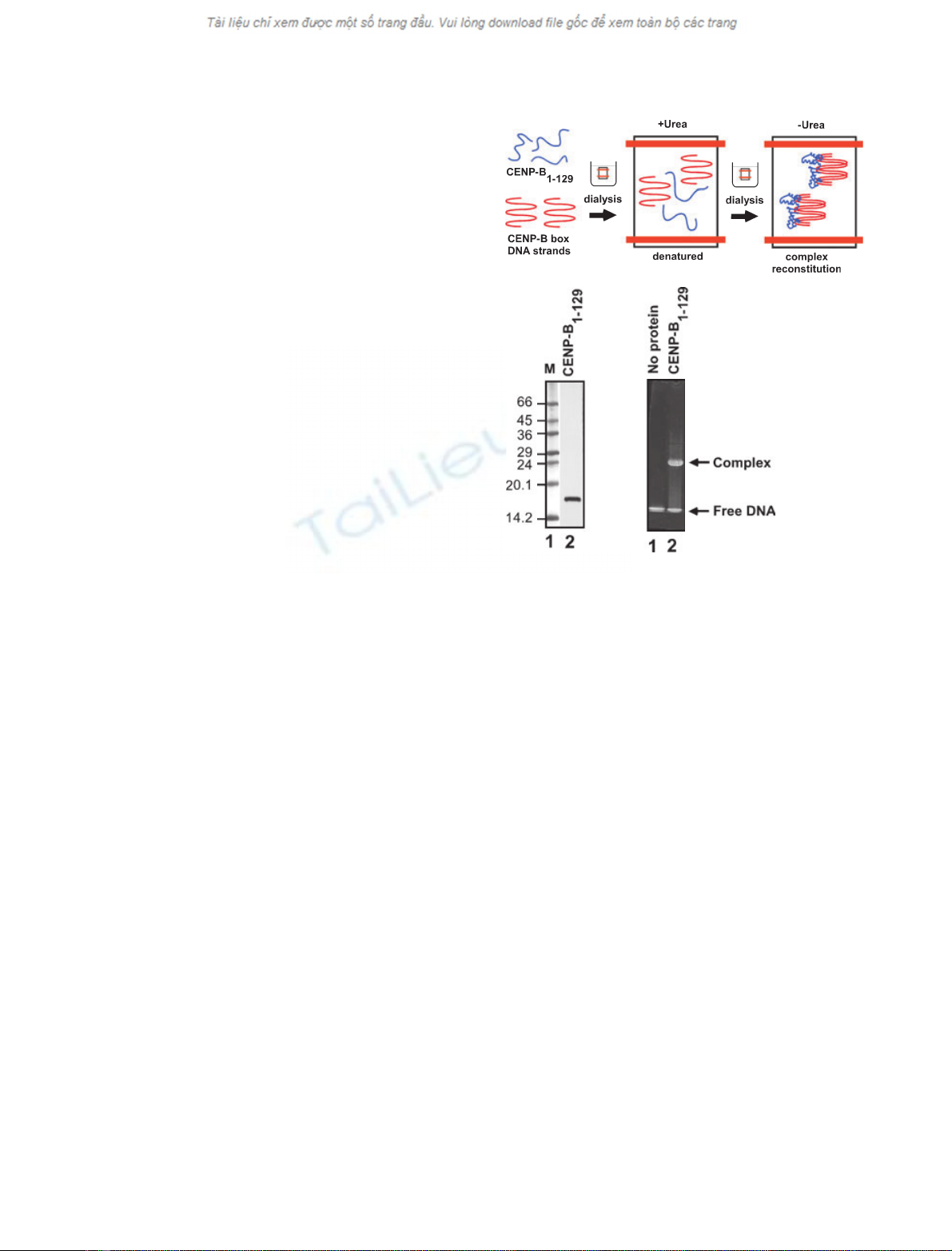
The complex-reconstitution assay
We tested whether the CpG methylations of the
CENP-B box DNA actually affect CENP-B binding
to the DNA, as suggested by the CENP-B(1–129)–
methylated DNA complex model. To do so, we
employed a complex-reconstitution assay with recom-
binant CENP-B(1–129) and the methylated and un-
methylated CENP-B box DNAs (21 bp; Fig. 2A). As
the recombinant CENP-B(1–129) was only detected in
the insoluble fraction when it was expressed in
Escherichia coli cells, CENP-B(1–129) was purified
under denaturing conditions in the presence of 6 m
urea (Fig. 2B). In the complex-reconstitution assay,
CENP-B(1–129) formed the complex with the CENP-
B box DNA during the refolding process by dialysis
against buffer without urea. The complex-formation
efficiency was monitored by a gel-shift assay
(Fig. 2C).
A
BC
Fig. 2. The complex-reconstitution assay with CENP-B(1–129) and
the CENP-B box DNA. (A) Schematic presentation of the complex-
reconstitution assay. CENP-B(1–129) and the CENP-B box DNA
strands are shown in blue and red, respectively. CENP-B(1–129)
and the CENP-B box DNA were mixed under denaturing conditions
in the presence of 6 Murea. The CENP-B(1–129)–DNA complex
was reconstituted during the refolding process by dialysis against
buffer without urea. (B) The purified recombinant CENP-B(1–129)
protein was analyzed by 15–25% SDS/PAGE. Lane 1 indicates
molecular mass markers (M), and lane 2 indicates CENP-B(1–129)
purified under denaturing conditions in the presence of 6 Murea.
(C) CENP-B(1–129)–DNA complex formation. The CENP-B(1–129)–
DNA complex, reconstituted by the method shown in panel A, was
analyzed by 20% nondenaturing PAGE. Bands were visualized by
ethidium bromide staining.
CpG methylation reduces human CENP-B binding
1Y. Tanaka et al.
284 FEBS Journal 272 (2005) 282–289 ª2004 FEBS
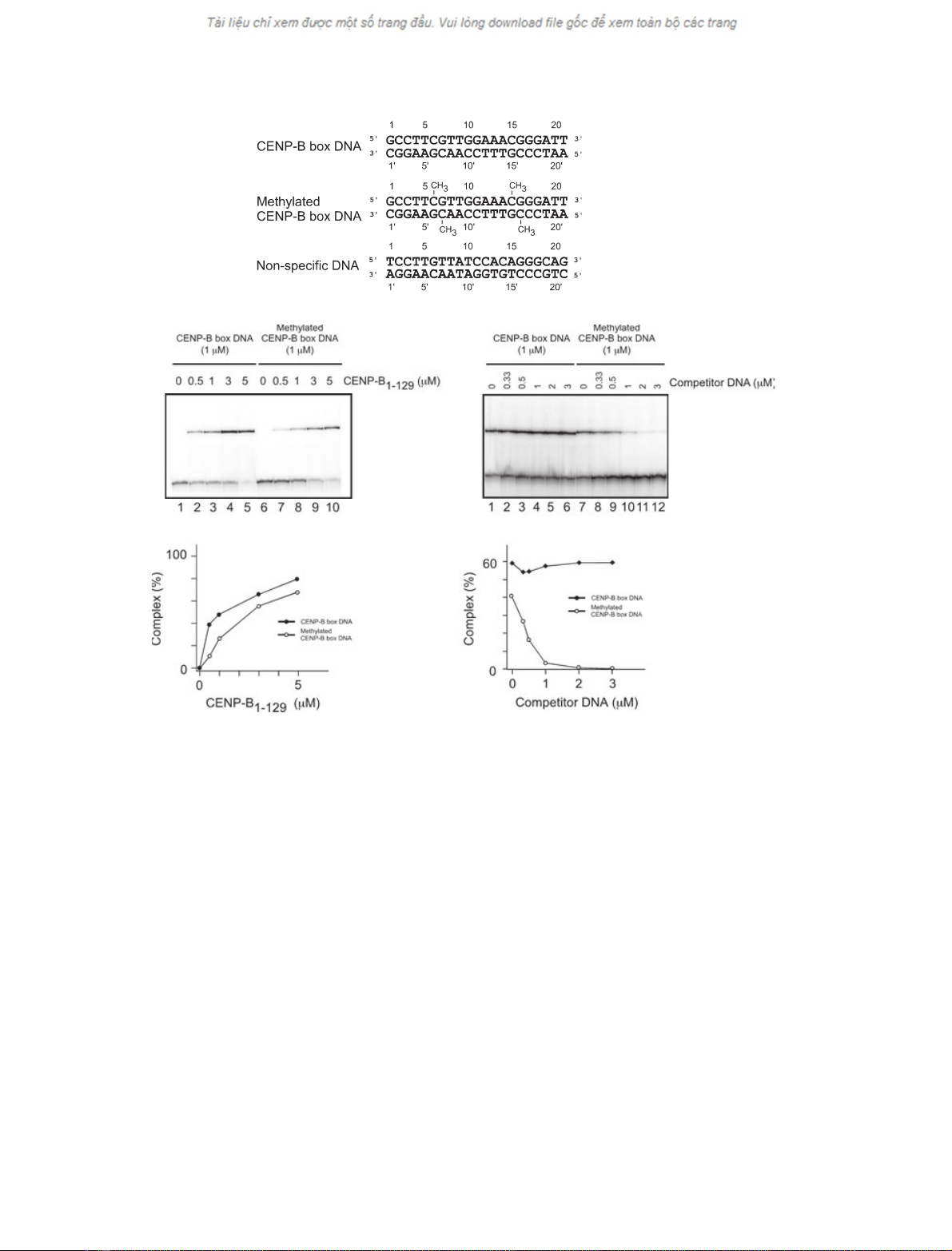
CENP-B(1–129) preferentially binds to the
unmethylated CENP-B box DNA rather than the
methylated form
Using the complex-reconstitution assay, we tested the
binding of CENP-B(1–129) to the unmethylated and
methylated CENP-B box DNAs (Fig. 3A). As shown
in Fig. 3B, CENP-B(1–129) efficiently formed the com-
plex with the unmethylated CENP-B box DNA in a
concentration-dependent manner (lanes 1–5). CENP-
B(1–129) also formed the complex with the methylated
CENP-B box DNA (Fig. 3B, lanes 6–10), but with
slightly reduced efficiency, as compared with the
unmethylated CENP-B box DNA (Fig. 3C). Therefore,
CENP-B has the potential to bind to the methylated
CENP-B box DNA.
A
B
CE
D
Fig. 3. CENP-B(1–129) preferentially binds to unmethylated CENP-B box DNA. (A) The 21-mer CENP-B box DNA and the 21-mer nonspecific
DNA (DnaA box DNA [30]), used in this study. The methylated cytosine residues in the methylated CENP-B box DNA are labeled by CH
3
in
the middle row. (B) Gel-shift analysis of complex formation between CENP-B(1–129) and the CENP-B box DNA, complexed with increasing
amounts of CENP-B(1–129), by 20% PAGE. The unmethylated CENP-B box DNA (lanes 1–5; 1 lM) and the methylated CENP-B box DNA
(lanes 6–10; 1 lM) were used in this study. The CENP-B(1–129) concentrations were 0 lM(lanes 1 and 6), 0.5 lM(lanes 2 and 7), 1 lM
(lanes 3 and 8), 3 lM(lanes 4 and 9), and 5 lM(lanes 5 and 10). (C) Graphic representation of the complex formation rates shown in panel
B. Unmethylated (d) and methylated (s) CENP-B box DNAs, respectively. (D) Competition analysis for CENP-B binding between the un-
methylated and methylated CENP-B box DNAs. The
32
P-labeled unmethylated CENP-B box DNA (1 lM)orthe
32
P-labeled methylated CENP-
B box DNA (1 lM) was mixed with the indicated amounts of the unlabeled competitor methylated or unmethylated CENP-B box DNAs,
respectively, in the presence of CENP-B(1–129) (3 lM). The complexes were analyzed by 20% PAGE. (E) Graphic representation of the com-
plex formation rates shown in panel D. Closed and open circles indicate experiments with the
32
P-labeled unmethylated (d) and methylated
(s) CENP-B box DNAs. Complex formation rates (%) are plotted against the concentrations of the competitor DNAs.
Y. Tanaka et al.CpG methylation reduces human CENP-B binding
1
FEBS Journal 272 (2005) 282–289 ª2004 FEBS 285
Next, we tested the binding affinity of CENP-B(1–
129) to the methylated CENP-B box DNA. To do so,
we performed a competition assay. In this assay,
complex formation between CENP-B(1–129) and the
32
P-labeled unmethylated or methylated CENP-B box
DNAs was tested in the presence of unlabeled compet-
itor DNA. As shown in Fig. 3D, complex formation
between CENP-B(1–129) and the
32
P-labeled methyla-
ted CENP-B box DNA was significantly inhibited in
the presence of only one-third of the amount of unlabe-
led unmethylated CENP-B box DNA (0.33 lm), relat-
ive to the
32
P-labeled methylated form (1 lm) (lanes
7–12, and E). In contrast, complex formation between
CENP-B(1–129) and the
32
P-labeled unmethylated
CENP-B box DNA was not affected, even in the pres-
ence of a threefold excess of unlabeled methylated
CENP-B box DNA (3 lm) (Fig. 3D, lanes 1–6, and E).
These results indicate that CENP-B preferentially
forms a complex with the unmethylated CENP-B box
DNA, rather than the methylated CENP-B box DNA.
The CENP-B binding affinity to the methylated
CENP-B box DNA is similar to the level of
nonspecific DNA binding
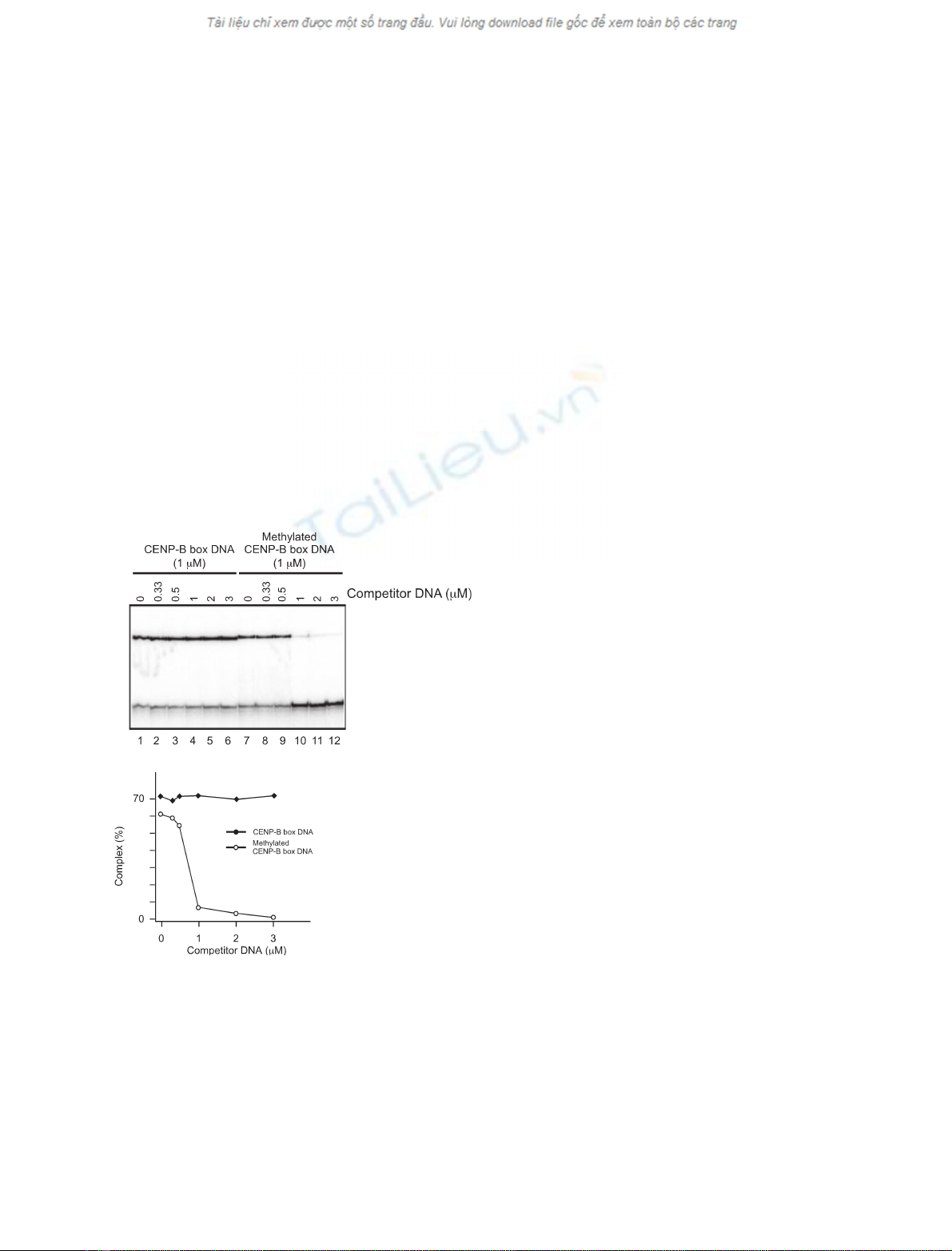
In order to compare the CENP-B binding to the
CENP-B box DNA and a nonspecific DNA, we per-
formed a competition assay with the methylated or
unmethylated CENP-B box DNAs and a nonspecific
DNA. The DnaA box sequence, which is not specific-
ally bound by CENP-B, was used as nonspecific DNA
(Fig. 3A, bottom row). The complex formation
between CENP-B(1–129) and the
32
P-labeled unmethy-
lated CENP-B box DNA was not affected when it was
titrated with unlabeled nonspecific DNA (Fig. 3A and
4A, lanes 1–6, and B). On the other hand, the complex
between CENP-B(1–129) and the
32
P-labeled methyla-
ted CENP-B box DNA was significantly dissociated in
the presence of an equal amount of unlabeled nonspe-
cific competitor DNA (1 lm) (Fig. 4A, lanes 7–12, and
B). Therefore, the CpG methylation at sites 1 and 3 of
the CENP-B box DNA reduces the CENP-B binding
affinity almost to the level of nonspecific DNA binding.
Discussion
In this study, we found that CpG methylation of the
CENP-B box sequence reduces the binding affinity
between CENP-B and the CENP-B box DNA nearly
to the level of nonspecific binding. In the CENP-B(1–
129)–methylated DNA complex model, the additional
methyl groups on cytosine 7¢and cytosine 15 caused
steric clashes with the side chains of Thr44 and
Arg125, respectively (Fig. 1C). In site 3, the CENP-B
a-helix 8 (120–129 amino acid residues), containing
four Arg residues (Arg125, 127, 128 and 129), penet-
rates perpendicularly into the major groove around the
CpG sequence. Interestingly, the Arg125 side chain
directly formed a hydrogen bond with the N7 atom of
guanine 16, and specifically recognized the site 3
sequence (Fig. 1C, right). In contrast, the Arg127, 128
and 129 residues bound to the backbone phosphates,
and did not directly interact with the DNA bases.
Steric hindrance of the specific interaction between
Arg125 and the N7 atom of guanine 16 by the CpG
methylation may be a reason for the reduced specificity
of CENP-B to the CENP-B box sequence.
What is the functional meaning of the CpG methyla-
tion of the CENP-B box DNA? Recently, a link
between centromeric heterochromatin formation and
the RNA interference (RNAi) machinery was discov-
ered. In fission yeast, the RNAi machinery is required
for chromosome segregation, gene silencing and nor-
mal centromere function [
326–28]. In chicken–human
A
B
Fig. 4. CpG methylation reduces the affinity between CENP-B and
the CENP-B box DNA to nearly the level of nonspecific DNA bind-
ing. (A) The
32
P-labeled CENP-B box DNA (1 lM) was mixed with
the indicated amounts of the unlabeled nonspecific DNA competitor
in the presence of CENP-B(1–129) (3 lM). The complexes were
analyzed by 20% PAGE. (B) Graphic representation of the complex
formation rates shown in panel A.
32
P-labeled unmethylated (d)
and methylated (s) CENP-B box DNAs. Formation rates (%) are
plotted against the concentrations of the competitor DNAs.
CpG methylation reduces human CENP-B binding
1Y. Tanaka et al.
286 FEBS Journal 272 (2005) 282–289 ª2004 FEBS


























