Isolation, enzymatic properties, and mode of action of an
exo-1,3-b-glucanase from
Trichoderma viride
Anna A. Kulminskaya
1
, Karl K. Thomsen
2,
*, Konstantin A. Shabalin
1
, Irina A. Sidorenko
1
, Elena V. Eneyskaya
1
,
Andrew N. Savel’ev
3
and Kirill N. Neustroev
1
1
Petersburg Nuclear Physics Institute, Russian Academy of Science, Russia;
2
Carlsberg Laboratory, Department of Physiology,
Copenhagen, Denmark;
3
St Petersburg Technical University, Biophysics Department, Russia
An exo-1,3-b-glucanase has been isolated from cultural
filtrate of Trichoderma viride AZ36. The N-terminal
sequence of the purified enzyme (m¼61 ^1kDa)
showed no significant homology to other known glucanases.
The 1,3-b-glucanase displayed high activity against
laminarins, curdlan, and 1,3-b-oligoglucosides, but acted
slowly on 1,3-1,4-b-oligoglucosides. No significant activity
was detected against high molecular mass 1,3-1,4-b-
glucans. The enzyme carried out hydrolysis with inversion
of the anomeric configuration. Whereas only glucose was
released from the nonreducing terminus during hydrolysis of
1,3-b-oligoglucosides, transient accumulation of gentiobiose
was observed during hydrolysis of laminarins. The
gentiobiose was subsequently degraded to glucose. The
Michaelis constants K
m
and V
max
have been determined for
the hydrolysis of 1,3-b-oligoglucosides with degrees of
polymerization ranging from 2 to 6. Based on these data,
binding affinities for subsites were calculated. Substrate
binding site contained at least five binding sites for sugar
residues.
Keywords:exo-1;3-b-glucanase; Trichoderma viride;
anomerity of hydrolysis.
Enzymes hydrolyzing 1,3-b-D-glucans occur in a variety of
organisms [1]. 1,3-b-Glucanases hydrolyze the O-glyco-
sidic linkages of 1,3-b-linked glucans and are classified
according to their mode of action. The exo-1,3-b-glucanases
(EC 3.2.1.58) sequentially release glucose residues from the
nonreducing terminus of a substrate while the endo-1,3-b-
glucanases (EC 3.2.1.39) are capable of cleaving internal
1,3-b-linkages at random sites along the polysaccharide
chain, releasing short oligosaccharides. 1,3-b-Glucanases
have been isolated from bacteria [2], yeast and fungi [3–5],
plants [6,7], and marine organisms [8,9]. It has been
suggested that plant 1,3-b-glucanases may protect the
germinating grain against pathogen attack [10]. Microbial
1,3-b-glucanases play an essential role in development and
differentiation of saprophyte and mycoparasite cultures
[11–13] while 1,3-b-glucanases from the filamentous fungi
Coprinas seem to be involved in the process of stipe
elongation [14]. In Saccharomyces cerevisiae the produc-
tion of exo-1,3-b-glucanases is growth-associated and cell-
cycle regulated, suggesting that their activities are required
at specific stages during morphogenesis [15,16]. Most
organisms synthesize multiple 1,3-b-glucanases rather than
a single enzyme [17] and complete degradation of 1,3-b-
glucans by fungi is often accomplished by synergistic action
of endo- and exo-glucanases [18]. These enzymes have
received attention in many fields of science and biotech-
nology because many cultures of microorganisms widely
used in industry produce 1,3-b-glucanases, which are essen-
tial for cell-cycle functions [19,20] and due to their increasing
importance in modification of b-glucans for pharmaceutical
purposes [21,22]. Despite a number of reports describing
exo-1,3-b-glucanases from different sources [19,20,23–25],
the subsite structure of the substrate binding site as well
as the affinity and the number of subsites have not been
analyzed for most of these enzymes. The present study
describes the isolation and characterization of an exo-1,3-b-
glucanase from the filamentous fungus Trichoderma viride
AZ36. The subsite structure was evaluated by steady-state
kinetics using 1,3-b-oligoglucosides with a different degree
of polymerization. The mode of action and specificity as
well as stereoselectivity of hydrolysis catalyzed by the exo-
1,3-b-glucanase were studied by NMR spectroscopy.
MATERIALS AND METHODS
Substrates
Laminarin from Laminaria digitata, barley 1,3-1,4-b-glucan,
lichenan from Cetraria islandica, gentiobiose, cellulose,
Correspondence to K. N. Neustroev, Petersburg Nuclear Physics
Institute, Gatchina, St Petersburg, 188350, Russia.
Fax: 1781271 32303, Tel.: 1781271 32014,
E-mail: neustk@omrb.pnpi.spb.ru
Enzymes: 1,3-b-glucanase, 1,3-b-D-glucan glucanohydrolase,
laminarinase (3.2.1.39); exo-b-1,3-glucanase, 1,3-b-D-glucan
glucohydrolase (EC 3.2.1.58); a-glucosidase (EC 3.2.1.20);
glucoamylase (EC 3.2.1.3); b-D-glucosidase, b-D-glucoside
glucohydrolase (EC 3.2.1.21).
Definition: G4G4G3G, b-D-Glcp-(1!4)-b-D-Glcp-(1!4)-b-
D-Glcp-(1!3)-b-D-Glcp; G4G3G, b-D-Glcp-(1!4)-b-D-Glcp-
(1!3)-b-D-Glcp; G3G3G3G3G, b-D-Glcp-(1!3)-b-D-Glcp-(1!3)-
b-D-Glcp-(1!3)-b-D-Glcp-b-D-Glcp-(1!3)-b-D-Glcp; G3G3G3G,
b-D-Glcp-(1!3)-b-D-Glcp-(1!3)-b-D-Glcp-(1 !3)-b-D-Glcp;
G3G3G, b-D-Glcp-(1!3)-b-D-Glcp-(1!3)-b-D-Glcp; G3G,
b-D-Glcp-(1!3)-b-D-Glcp; G6G, gentiobiose.
*Present address: Fussingsvej 8, I, DK-8700 Horsens, Denmark.
(Received 3 July 2001, revised 20 September 2001, accepted
27 September 2001)
Abbreviations: DP, degree of polymerization; PHMB,
p-hydroxymercuribenzoic acid sodium salt.
Eur. J. Biochem. 268, 6123–6131 (2001) qFEBS 2001

cellobiose, p-nitrophenyl cellotrioside, p-nitrophenyl
b-D-glucopyranoside were from Sigma Chemical Co. (St
Louis, MO, USA). Curdlan from Alcaligenes faecalis and
pustulan from Umbilicaria popullosa were kindly donated
by I. J.Goldstein (Michigan University, USA). Laminarin
from Laminaria cichorioides was kindly donated by A. V.
Kir’yanov (Institute of Organic Chemistry, Moscow,
Russia). p-Hydroxymercuribenzoic acid sodium salt
(PHMB) was from Merck (Germany).
Mixed linkage oligosaccharides (for oligosaccharide
definitions, see footnotes): G4G4G3G and G4G3G were
produced by digestion of barley glucan with a 1,3-1,4-b-
glucanase and purified [26].
The G3G3G3G3G3G, G3G3G3G3G, and G3G3G3G were
prepared by formic acid hydrolysis of curdlan followed by
purification of 1,3-b-oligoglucosides [26,27]. G3G and
G3G3G were produced by digestion of laminarin with a
commercially available laminarinase from Trichoderma sp.
purchased from Sigma Chemical Co. Laminarin (80 mg)
was dissolved in 2 mL of 20 mMsodium acetate buffer,
pH 5.0, and digested using <0.005 units of the enzyme
per mg of laminarin. Incubation was at 37 8Cfor
60 min. The reaction mixture was fractionated on a
Sephadex G-25 (Fine) column equilibrated in water,
separating oligosaccharides of degree of polymerization
(DP) 2– 6 from the high molecular mass fraction. Following
freeze drying the 1,3-b-oligoglucosides were fractionated
on a TSK NH
2
-60 column (5 mm, 4.6 250 mm) from
Pharmacia Biotech (Uppsala, Sweden) in 80% acetonitrile
in water (v/v).
The purity of b-oligoglucosides was analyzed by TLC
and
1
H and
13
C NMR spectroscopy as described below.
Published values for
1
Hand
13
C chemical shifts of b-oligo-
saccharides with different DP and linkage types [26,28–30]
were used for structure determination of the obtained
compounds.
General methods
SDS/PAGE was carried out according to Laemmli [31] and
isoelectric focusing was on Servalyt PRECOATES plates
3– 10 (Serva Electrophoresis GmbH, Heidelberg, Germany).
Protein concentration was measured following the Lowry
procedure using BSA as a standard [32].
1
H-NMR spectra
and
13
C-NMR spectra were recorded with an AMX-500
Bruker spectrometer. Prior to NMR analysis laminarin,
buffer components, and the enzyme were freeze-dried twice
from D
2
O. The measurements were made in 20 mMsodium
phosphate buffer (pD 6.0) at room temperature and 50 mM
4,4-dimethyl-4-silapentane sodium sulfonate was used as an
internal standard in 0.5 mL 99.8% D
2
O. The solvent reson-
ance was presaturated for 0.5 s with a decoupler operating at
24 dB in the CW mode. Data were acquired after a 758pulse
into 16K points, with a spectral width of 10 kHz and 2053
scans, including first five dummy scans. The spectra were
Lorentz-broadened by 1 Hz. The 4,4-dimethyl-4-silapen-
tane sodium sulfonate signal was used for adjustment of
phase and amplitude parameters in order to obtain correct
differential spectra. Oligosaccharide substrates and products
of enzymatic hydrolysis were analyzed by TLC on
Kieselgel 60 plates from Merck (Darmstadt, Germany)
with a mobile phase of ethyl acetate/acetic acid/water
(2 : 1 : 1). Plates were developed at room temperature, air
dried and sprayed with 5% H
2
SO
4
in 1-propanol followed by
incubation at 120 8C for 8 min. N-Terminal amino-acid
sequencing was conducted using the Edman degradation and
phenylisothiocyanate amino-acid analysis. The Procise
Protein Sequencing System (Applied Biosystems, Foster
City, California 94404) was employed.
Growth conditions
The T. viride AZ36 from Petersburg Nuclear Physics Insti-
tute strain collection was grown at 30 ^18C for 72 h in a
10-L fermentor with constant stirring. The growth medium
contained (g per L) KH
2
PO
4
, 1; NaNO
3
, 1.5; (NH
4
)
2
SO
4
,
1.5; MgSO
4
7H
2
), 0.5; wheat bran, 40.
Purification of the exo-1,3-b-glucanase
All steps were carried out at 4 8C. Mycelium was removed
by centrifugation (3000 g, 30 min), and the supernatant was
concentrated 20-fold by use of hollow fibers with a nominal
molecular mass limit of 25 kDa (‘Kirishi’, Kirishi, Russia).
During the process the buffer was changed to 20 mM
Tris/HCl, pH 7.5 (buffer A). The crude 1,3-b-glucanase
preparation was loaded on a DEAE-Sephadex column
(50 200 mm) equilibrated with buffer A and bound pro-
tein was eluted with 1 MNaCl in the same buffer. Following
concentration to 25 mL on an Amicon PM-30 membrane
and dialysis against buffer A, the fraction was loaded onto a
TSK column (21.5 150 mm) equilibrated with buffer
A. Elution was performed by using a linear 0– 0.5 MNaCl
gradient in buffer A. Fractions containing exo-1,3-b-
glucanase activity were concentrated to 4 mL on an Amicon
PM-30 membrane, dialyzed against 20 mMTris/HCl,
pH 7.2 (buffer B) and chromatographed on a Mono Q HR
(5/5) (Pharmacia, Sweden). Bound protein was eluted by
applying a linear 0–0.5 MNaCl gradient in buffer B.
Finally, the exo-1,3-b-glucanase preparation was concen-
trated to 3 mL using an Amicon PM-30 membrane, dialysed
against 20 mMsodium acetate buffer, pH 5.0 (buffer C).
Saturated (NH
4
)
2
SO
4
in buffer C was added to a final
concentration of 1.7 Mbefore applying the enzyme prepar-
ation onto a phenyl-Superose HR (5/5) column (Pharmacia,
Sweden) equilibrated with 1.7 M(NH
4
)
2
SO
4
in buffer C.
Bound protein was eluted by using a linear gradient
(0–100%) of 20 mMsodium acetate buffer, pH 5.0, and
fractions with the exo-1,3-b-glucanase activity were
dialyzed against buffer C, then against deionized water,
and freeze dried.
Enzyme assays
Exo-1,3-b-glucanase activity was analyzed by measuring
the amount of glucose released from laminarin. Standard
assays (0.25 mL) were in 20 mMsodium acetate buffer,
pH 4.5, and contained 0.5 mg of laminarin and crude or
purified enzyme extract corresponding to at least 0.2 mg
of pure exo-1,3-b-glucanase. Incubation was at 37 8C for
10–30 min and the reaction was stopped by boiling for
5 min. Glucose formation was measured by the glucose
oxidase method [33]. Alternatively, enzyme activity was
evaluated by measuring the formation of reducing sugars
according to the Somogyi-Nelson method [34]. One unit of
the enzyme activity produced 1 mmol of glucose:min
21
at
6124 A. A. Kulminskaya et al. (Eur. J. Biochem. 268)qFEBS 2001
pH 4.5, 37 8C, with laminarin as substrate. b-Glucosidase
activity was measured using p-nitrophenyl b-D-glucopyr-
anoside as substrate according to [35].
The influence of pH and temperature on activity of the
enzyme was studied by the glucose oxidase method. The
effect of pH on activity was measured at 37 8C for 10 min in
the range pH 3– 9 in 100 mMsodium phosphate/citrate
buffers. Reaction mixture: 400 mL of buffer and 10 mLof
enzyme (0.01 U) in 2 mMsodium acetate buffer, pH 4.5,
was mixed with 50 mL of laminarin (20 mg:mL
21
in water)
or with 20 mL of G3G3G3G (40 mMin water). The
influence of pH on stability of the enzyme was studied by
incubating samples of the enzyme at 20 8C for 16 h in
100 mMsodium phosphate/citrate buffers ranging from
pH 3 to pH 9, followed by measuring the residual activity of
the enzyme using standard conditions.
The effect of temperature on activity was measured at
pH 4.5 in 100 mMsodium citrate buffer over a temperature
range of 25–80 8C. The reaction mixture (0.4 mL) con-
taining 1 mg of laminarin was preincubated at various
temperatures in the range mentioned above. Then 50 mL
(0.005 U) of enzyme solution in the same buffer was added
and incubated for 10 min. The reaction was stopped by
addition of 1 mL of copper reagent (Somogyi reagent) and
boiling for 5 min. Liberated glucose was measured by the
Somogyi-Nelson method [34]. To study stability in relation
to temperature, the purified exo-1,3-b-glucanase was
incubated at various temperatures for 10 min, in 100 mM
sodium citrate buffer, pH 4.5. After cooling, the residual
activity was determined by the glucose-oxidase method as
described above.
Effect of metal ions on activity was determined in 50 mM
sodium acetate buffer, pH 4.5, at 37 8C after 15 min of
incubation with different ions.
Kinetic parameters
TheMichaelis–MentenconstantsK
m
and V
max
were deter-
mined from the Lineweaver–Burk representation of data
obtained by measuring the initial rate of substrate hydrolysis.
The purified exo-1,3-b-glucanase (0.01–1 U) was incubated
with different substrates in 0.5 mL of 20 mMsodium acetate
buffer, pH 4.5. After the reaction was stopped by boiling,
liberated glucose was measured as described above.
Concentrations of laminarins from 1 to 0.05 mg:mL
21
,
curdlan concentrations from 15 to 1 mg:mL
21
,and
b-oligoglucoside concentrations from 12 to 0.1 mMwere
used.
Enzyme specificity
The specificity of the exo-1,3-b-glucanase was studied by
analyzing the products of hydrolysis of different substrates.
Laminarins with different degrees of ramification as well as
curdlan, pustulan, and b-oligoglucosides differing in DP and
linkage type were used. Hydrolysis was stopped after
appropriate time intervals by boiling for 5 min. The
resulting products were analysed qualitatively by TLC and
quantitatively by HPLC using a TSK-NH
2
-60 column
(5 mm, 4.6 250 mm) from Pharmacia Biotech (Uppsala,
Sweden) in 80% acetonitrile in water (v/v) with refracto-
metric detection. Glucose and 1,3-b-oligoglucosides of DP
2–6 were used as standards. To investigate activity of the
exo-1,3-b-glucanase towards different soluble and insoluble
b-glucans, substrates at a concentration of 5 mg:mL
21
were
incubated with 2–3 U of enzyme at 37 8C for various time
intervals.
For studies of the mode of action in the hydroysis of
laminarins, intermediate products were fractionated by
HPLC on a Dextro-Pak cartrige column (8 10 mm)
WATO85650 from Millipore-Waters (Bedford, MA, USA)
using isocratic elution with water. The purified products of
hydrolysis were analyzed by
1
H and
13
C NMR spectroscopy
as described previously [30].
Determination of the stereochemical course of hydrolysis
The enzyme was dissolved in 20 mMsodium phosphate
buffer, pH 6.0, made up in D
2
O. The reaction mixture of
0.6 mL contained 20 mg:mL
21
laminarin in sodium
phosphate buffer. All NMR measurements were performed
at ambient temperature (<17 8C). After accumulation of the
initial spectrum, 80 U of the exo-1,3-b-glucanase was added
and the reaction mixture was brought to 37 8C within 5 min.
The stereochemical course of hydrolysis was monitored by
collecting spectra at 6, 20, and 40 min after addition of the
enzyme. Direct
1
H NMR analysis of the exo-1,3-b-glucanase
action on 1,3-b-oligoglucosides was carried out at a substrate
concentration of <10 mMin 0.6 mL of the same buffer.
RESULTS AND DISCUSSION
Enzyme purification
The T. viride AZ36 used in this study is a producer of
mainly exo-1,3-b-glucanase. An exo-1,3-b-glucanase was
purified 25-fold from concentrated culture supernatants of
T. viride in four chromatographic steps. About 40% of
the initial activity was recovered and the purified enzyme
Table 1. Purification of the exo-1,3-b-glucanase.
Purification step
Volume
(mL)
Total protein
(mg)
Total activity
(U)
Specific activity
(U:mg
21
)
Purification
(fold)
Yield
(%)
Cultural liquid 1000 250 630 2.5 1 100
Ultrafiltration 50 125 570 4.6 1.8 90
DEAE-Sephadex chromatography 50 70 440 6.3 2.5 70
DEAE-5PW chromatography 25 15 315 21 8.4 50
Mono Q chromatography 4 6 283 47 18.8 45
Phenyl Superose chromatography 3 4 252 63 25.2 40
qFEBS 2001 Exo-1,3-b-glucanase from T. viride (Eur. J. Biochem. 268) 6125
had a specific activity of 63 U:mg
21
(Table 1). In SDS/
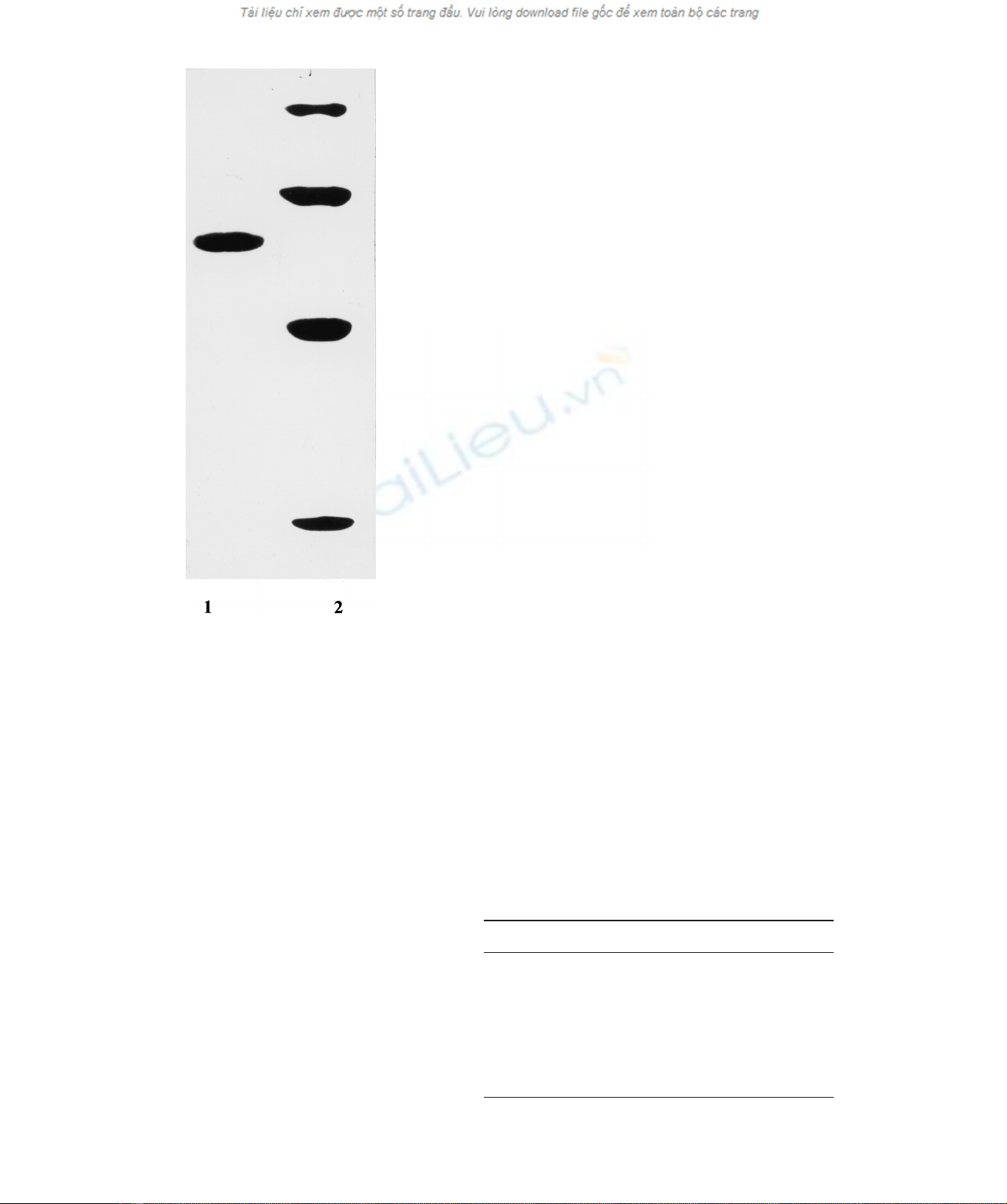
PAGE analysis the protein appeared as a single polypeptide
with an apparent molecular mass of 61 ^1 kDa (Fig. 1).
Small amounts of endo-1,3-b-glucanase were separated
from the main fraction during the first steps of purification.
During the purification process, the exo-1,3-b-glucanase
and b-glucosidase activities were efficiently separated, and
the purified exo-1,3-b-glucanase preparation possessed
less than 0.1% b-glucosidase activity. Analytical gel
filtration on a Superose 12 column demonstrated that the
exo-1,3-b-glucanase had an apparent molecular mass of
60 ^5 kDa suggesting that the enzyme was a monomer
(data not shown).
Multiple forms of Trichoderma harzianum exo-1,3-b-
glucanases with a wide range of molecular masses up to
35 kDa have been reported [13]. The multiplicity of
secreted forms of the Trichoderma enzymes might be
caused by postsecretory proteolysis by acid proteases also
secreted by the fungus [36,37]. The strain T. viride AZ36
employed in the present study has a lower level of secreted
proteases than the other Trichoderma species (data not
shown), which is likely to be one of the reasons that a
homogenous exo-1,3-b-glucanase preparation could be
obtained.
General properties
The purified exo-1,3-b-glucanase displayed optimal activity
in hydrolysis of laminarin from L. digitata and 1,3-b-
oligoglucosides at pH 4.5. The optimal temperature was
55 8C. Activity decreased rapidly at temperatures above
60 8C. At room temperature the enzyme was stable for 24 h
in the pH range 3.5 to 7.5. The isolelectric point was
4.2 ^0.05 and the N-terminal amino-acid sequence was:
AVDDFAPNTKQTPIALNNVLL. There are a number of
reports providing amino-acid sequence data for fungal exo-
1,3-b-glucanases such as Cochliobolus carbonum [38],
yeast C. albicans [39], Saccharomyces cerevisiae [40],
Yarrowia lipolitica [41] but none of these display significant
homology in their N-terminal amino-acid sequence to the
exo-1,3-b-glucanase described in here.
Whereas some divalent metal ions as well as EDTA had a
slight inhibitory effect on the exo-1,3-b-glucanase, MnSO
4
and MnCl
2
increased the activity towards laminarin
(Table 2). Similar dependence of the enzymic activity on
metal ions was reported for endo-1,6-a-mannanase from a
soil bacterium [42]. Based on the observation that neither
Hg
21
nor PHMB caused any inhibition of enzyme activity,
it is assumed that no essential SH-groups are involved as
functional groups. Most exo-1,3-b-glucanases of fungal
origin typically display optimal enzymatic activity in the pH
range between 4 and 6 [19,23,43]. Similarly the enzyme
from T. viride AZ36 displayed optimal activity at pH 4.5.
The stability in relation to pH and temperature was also
similar to that of other fungal exo- and endo-glucanases
[11], and like most exo-1,3-b-glucanases that have been
studied, the enzyme described here was not inhibited by
heavy metal ions and PHMB.
The mode of action of the exo-1,3-b-glucanase was
analyzed employing 1,3-b-oligoglucosides with DP 3–6
and curdlan from A. faecalis [26] as substrates. Using TLC
and HPLC analyses it was demonstrated that glucose was
the only product formed during the enzyme-catalysed
reaction.
1
H NMR techniques was also used to study the
mode of action of purified exo-1,3-b-glucanase on curdlan
and reduced G3G3G3G3G. A signal at d¼5.248 p.p.m.
corresponding to reducing ends [26] did not increase during
hydrolysis. This observation unequivocally demonstrates
the absence of endo-type hydrolysis. Consequently, this
glucanase should be classified as an exo-glucohydrolase
(Fig. 2). The enzyme displayed only very low activity
against 1,3-1,4-b-glucans, lichenan, and barley glucan, and
Fig. 1. SDS/PAGE of the exo-1,3-b-glucanase from T. viride. Lane
1, purified exo-1,3-b-glucanase from T. viride;lane2,protein
standards – phosphorylase a(98 kDa), BSA (67 kDa), ovalbumin
(45 kDa), a-chymotrypsin (25 kDa).
Table 2. Effect of metal ions, PHMB, and EDTA on the exo-1,3-b-
glucanase activity towards laminarin.
Effector (1 mM) Relative activity (%)
None 100
Ca
21
97
Mn
21
136
Mg
21
89
Zn
21
83
Hg
21
86
PHMB 90
EDTA 81
6126 A. A. Kulminskaya et al. (Eur. J. Biochem. 268)qFEBS 2001
no activity was detected against 1,4-b-glucan, 1,4-b-oligo-
glucosides, pustullan (1,6-b-glucan), cellulose, and p-nitro-
phenyl b-glucoside. Whereas the K
m
values for mixed
linked 1,3-1,4-b-oligoglucosides and the corresponding
1,3-b-oligoglucosides were similar (Tables 3 and 4), the
V
max
value for hydrolysis of G4G3G was <260-fold lower
than for G3G3G. For the corresponding tetraoses the
difference was <4000-fold.
The mode of action of the T. viride exo-1,3-b-glucanase
in hydrolysis of 1,3-b-oligoglucosides and curdlan is typical
of fungal exo-1,3-b-glucanases [17,19,23]. The enzyme also
displayed some activity towards 1,3-1,4-b-oligoglucosides.
In contrast to barley exo-1,3-b-glucanase [44] and exo-
1,3-b-glucanase from C. albicans [45], the enzyme was
inactive against cellooligosaccharides and p-nitrophenyl
b-glucopyranoside.
Subsite stucture of the active center
For determination of the active center subsite structure the
kinetic parameters K
m
and V
max
of the hydrolytic reaction
were measured using 1,3-b-oligoglucosides of DP 2–6 as
substrates. The initial rate of hydrolysis was measured as a
function of substrate concentration. For all 1,3-b-oligo-
glucosides the reaction follows Michaelis–Menten kinetics.
Values for K
m
,V
max
and V
max
/K
m
for different substrates
shown in Table 4 indicate that oligoglucosides with lower
DP have lower V
max
. Because this enzyme exclusively
displays exo-activity in the hydrolysis of 1,3-b-oligogluco-
sides, the subsite theory of Hiromi [46,47] was employed for
construction of a subsite map for the estimation of affinities
and number of subsites in the active center of the enzyme.
According to this theory the kinetic parameters can be
expressed in a unified way in terms of subsite affinities A
i
of
msubsites and the intrinsic rate constant K
int
for glucosyl
bond cleavage [46]. As shown in [47], the subsite affinities
for sites A
225
of the exo-1,3-b-glucanase according to
Davies et al. numbering system for exo-glycanases [48]can
be calculated from the Eqn (1):
lnðVmax/KmÞn112lnðVmax/KmÞn¼2ðAn11/RTÞð1Þ
where K
m
and V
max
are the kinetic parameters of the exo-1,3-
b-glucanase-catalyzed hydrolysis of 1,3-b-oligoglucosides of
DP 3–6. Assuming that there is only one nonproductive
complex A
21
can be obtained from the Hiromi dependence
of 1/V
max
on exp[–A
n11
/(RT )] [47]. The value for A
1
can be
estimated from the Eqn (2) based on preliminary calculated
affinities A
2
–A
5
:
ðKmÞn¼55
exp ÿX
nÿ1
i¼ÿ1
Ai
RT
1exp ÿX
n
i¼1
Ai
RT
1…
!
ð2Þ
The calculated affinities A
21
and A
1
–A
5
are represented in
Fig. 3A. The diagram shows that negative values for binding
energy were at subsites 21to14, so that the exo-1,3-b-
glucanase from T. viride strain AZ336 possesses at least
five binding subsites of monomeric units at its binding site. A
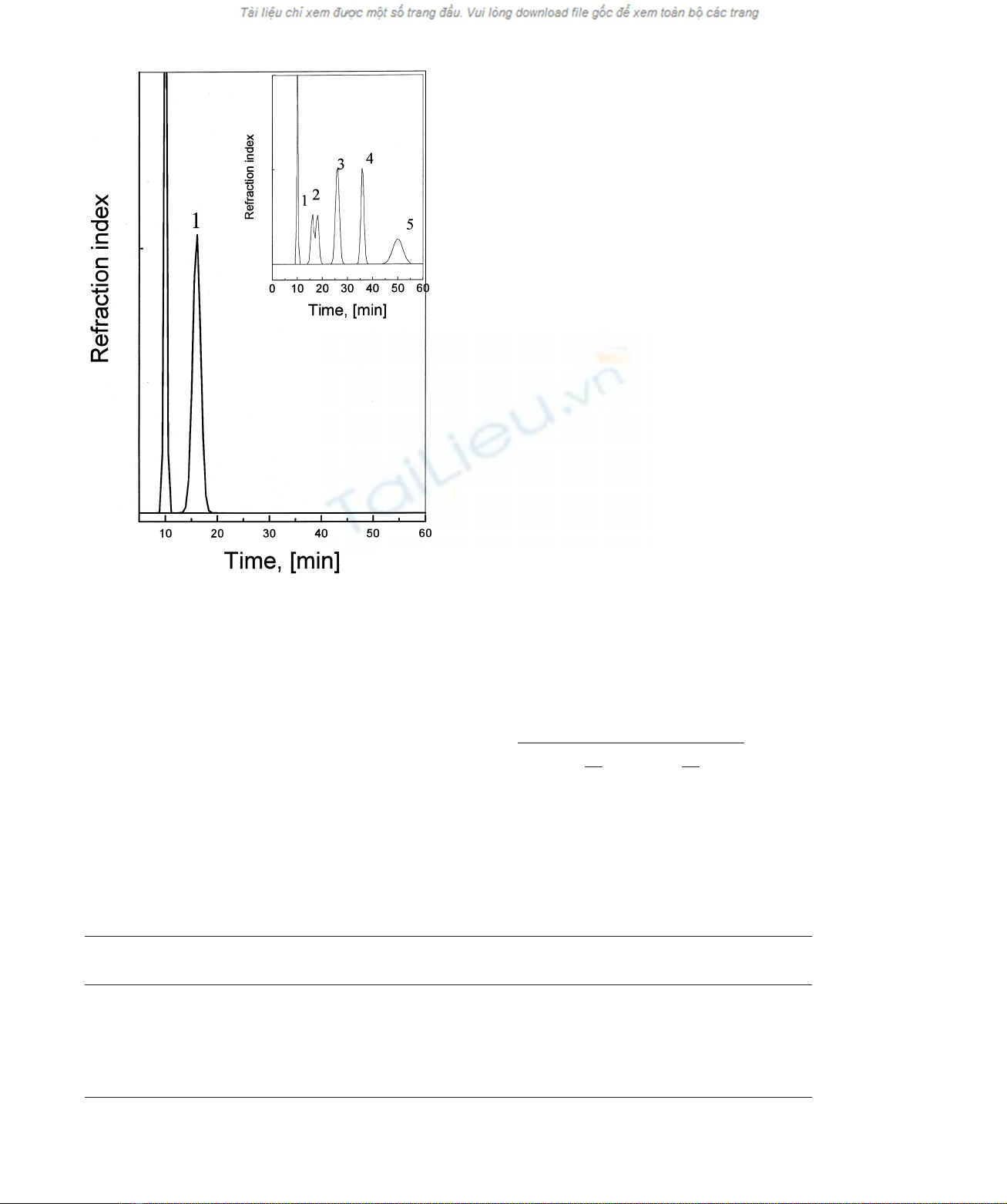
Fig. 2. HPLC analysis of the hydrolytic products resulting from the
exo-1,3-b-glucanase action on curdlan. Separation was performed on
a Lichrosorb NH
2
-60 column in 80% acetonitrile in water. Elution rate
was 1 mL:min
21
: 1, glucose. Inset: elution profile for a Lichrosorb
NH
2
-60 column separation of a mixture of 1,3-b-oligoglucosides:
1, glucose; 2, laminariobiose; 3, laminariotriose; 4, laminariotetraose;
5, laminariopentaose.
Table 3. Relative rate of hydrolysis of
b
-glucans, and oligosaccharides by the exo-1,3-b-glucanase.
Substrate
K
m
(mM)
10
3
:V
max
(mmol:min
21
:mg
21
)
Laminarin from L. digitata 0.12 mg:mL
21
17
Laminarin from Laminaria cichorioides 0.14 mg:mL
21
38
Curdlan 12 mg:mL
21
8.3
G4G3G 6.06 0.021
G4G4G3G 0.9 0.0034
G6G 2.5 0.185
qFEBS 2001 Exo-1,3-b-glucanase from T. viride (Eur. J. Biochem. 268) 6127


























