
Two CCAAT/enhancer binding protein sites in the cytochrome
P4503A1 locus
Potencial role in the glucocorticoid response
Elsa Rodrigues
1
, Marie-Jose
´Vilarem
2
, Vera Ribeiro
1,
*, Patrick Maurel
2
and Maria C. Lechner
1
1
Molecular Biology Unit, Faculty of Pharmacy, University of Lisbon, Portugal;
2
Unite
´128 INSERM, Montpellier, France
Induction of CYP3A genes by the ligand-activated preg-
nane-X-receptor (PXR) involves the interaction of other as
yet unidentified liver transcription factors. Here we show
that the CYP3A1 promoter contains two active sites con-
trolled by the CCAAT/enhancer-binding protein a(C/
EBPa), previously shown to regulate a number of liver stress
response genes. We have identified two functional C/EBP
binding sites at the CYP3A1 promoter that confer luciferase
activity to C/EBPacotransfected CHO cells. When inserted
upstream of a thymidine kinase promoter, oligonucleotides
corresponding to these elements ()350/)311 and )628/
)608), increase reporter gene expression when cotransfected
with a C/EBPaexpression vector. Point mutations in the
most conserved nucleotides in either element prevent binding
of C/EBPaand abolish transactivation of the CYP3A1
promoter. Moreover, we demonstrate that C/EBPaaccu-
mulates in the rat liver nuclei in response to dexamethasone,
and that under these conditions C/EBPabinds to the
CYP3A1 promoter elements. Our results suggest a correla-
tion between transcription of C/EBPa, nuclear protein
function and induction of CYP3A1 by dexamethasone in the
liver. They also support the notion that C/EBPaparticipates
in the up-regulation of the CYP3A1 gene in response to
synthetic glucocorticoids.
Keywords: cytochrome P450; CYP3A1 locus; C/EBP; regu-
latory elements; glucocorticoid induction.
Mammalian hepatic phenotypes are controlled through the
concerted action of a number of liver-enriched transcription
factors that act in cooperation with a number of ubiquitous
and ligand-activated factors to direct the selective expression
of liver-specific genes. The activity of each target gene is
modulated by dynamic arrays of counterpart transcription
factors that act upon liver differentiation and that are
stimulated by endogenous as well as by exogenous ligands [1].
Cytochrome P450 genes, members of the CYP1–4 fami-
lies, are abundantly expressed in the liver and are subjected
to complex regulatory networks that depend markedly on
the ontological development, and on the hormonal and
nutritional status of the animals [2]. Moreover, P450 mono-
oxygenases, as major body interfaces, are adaptive enzymes
highly responsive to induction and repression by environ-
mental and xenobiotic agents in general.
Recently, important advances have been made in under-
standing the mechanism of action of prototype inducers that
control the expression of hepatic P450 enzymes, namely
those mediated by the nuclear receptors AhR, CAR, PPAR
and PXR (reviewed, [3]). However, the role of these ligand-
activated receptors can be enhanced, reduced or inhibited by
the availability of other transcription factors that participate
in the organization of the transcription initiation complexes
in the native DNA context of each CYP locus. Such func-
tional interactions have not been fully defined, but their elu-
cidation is essential to understand the molecular basis of the
marked physiological variations often observed in the in vivo
response of each CYP gene to common inducing agents.
CYP3A1 has been widely investigated as a model of a
xenobiotic up-regulated liver expressed gene. Transcription
of CYP3A1 is virtually undetected in the adult rat liver, but
markedly induced by structurally diverse agents, including
the synthetic glucocorticoid agonist dexamethasone [4] and
the antagonist pregnenolone 16a-carbonitrile [5]. Such
paradoxical behaviour of the rat CYP3A1, and of other
orthologous genes, was shown to involve the pregnane X
receptor (PXR) [6–8], a member of the steroid hormone
receptor family. The mechanism of the glucocorticoid-
mediated activation of CYP3A1 relies on a dexamethasone
responsive unit in the promoter region of this gene [9,10].
This unit comprises elements that are targeted by the
activated PXR, as well as by COUP-TFs and HNF4 [11,12].
However, in vitro experiments have shown that mouse [6],
Correspondence to M. C. Lechner, Molecular Biology Unit,
Faculty of Pharmacy, University of Lisbon,
Av. Prof Gama Pinto, 1649–003 Lisbon, Portugal.
Fax: + 351 21 7946491, Tel.: + 351 21 7946490,
E-mail: clechner@ff.ul.pt
Abbreviations: C/EBP, CCAAT/enhancer binding protein; C/EBP
cons, C/EBP consensus oligonucleotide; CYP, cytochrome P450;
PXR, pregnane-X-receptor; AhR, aryl hydrocarbon receptor; CAR,
constitutive androstane receptor; PPAR, peroxisome proliferator-
activated receptor; RXR, retinoid-X-receptor; COUP-TF, chicken
ovalbumin upstream promoter transcription factor; HNF4, hepato-
cyte nuclear factor 4; TK, thymidine kinase; EMSA, electrophoretic
mobility-shift assay.
*Present address: Biochemistry Laboratory, Chemistry Departmental
Area, Faculty of Sciences and Technology, University of Algarve,
Campus de Gambelas 8000-117 Faro, Portugal.
(Received 30 September 2002, revised 14 November 2002,
accepted 5 December 2002)
Eur. J. Biochem. 270, 556–564 (2003) FEBS 2003 doi:10.1046/j.1432-1033.2003.03413.x

rat [13] and human [8,14] PXR receptors are moderately
activated by dexamethasone. This result is in contrast with
the strong in vivo transcriptional induction by this gluco-
corticoid [5,15–17], and suggests that PXR does not fully
explain the pattern of CYP3A1 induction. The importance
of other transcription factors for the positive regulation by
dexamethasone is also suggested by the observation that full
CYP3A1 induction can not be achieved in CV1 cells, that
lack liver-enriched transcription factors presumably needed
to elicit the response [18].
Other factors structurally unrelated to PXR have been
demonstrated to activate transcription of liver-expressed
genes, namely as potentiators of glucocorticoid responses.
CCAAT/enhancer-binding proteins (C/EBP) were shown to
play a role in adipocyte differentiation, as well as in the liver
acute-phase response (review, [19]). These factors create
multiple possibilities of combinatorial gene regulation in the
liver, by strategies that involve heterodimerization with
other b-zip proteins, and cross talk interactions with other
factors, such as the glucocorticoid receptor [20]. P450 genes,
namely Cyp2D5 [21,22], CYP2B [23–25], CYP2C12 [26] and
human CYP3A4 [27], have been shown to be the targets of
C/EBP factors in liver cells.
In the present work we investigated the presence of cis-
acting elements in the CYP3A1 promoter. Dynamic trans-
fection–transactivation assays were performed to evaluate
the potential activity of recombinant C/EBPaon the previ-
ously cloned CYP3A1 promoter region [28]. Two C/EBP
binding sites were identified by proteinÆDNA gel mobility-
shift assays. To assess the biological significance of these
regulatory elements in the response to glucocorticoids, the
concurrence of the CYP3A1 and the C/EBP responsiveness
was investigated in the rat liver upon synthetic glucocorticoid
administration. The hypothesis of a cause–effect relationship
is suggested by the time-course analysis of C/EBPaand
CYP3A1 mRNA and protein accumulation in the liver.
Our in vitro and the in vivo results indicate that C/EBPais
an activator of the CYP3A1 gene promoter and suggest a role
for this liver enriched transcription factor in the transcrip-
tional activation of CYP3A1 by synthetic glucocorticoids.
Materials and methods
Animals and treatments
Male adult Wistar rats, bred at the Gulbenkian Institute
animal house, Ociras, Portugal, were used in this investiga-
tion. Rats were maintained with standard chow and water
ad libitum, until 24 h before hormone treatment. Dexameth-
asone 21-phosphate (Sigma) was given intragastrically, in
aqueous solution, in a dose of 40 mg per kg body mass, and
the animals were sacrificed at different time points after drug
administration. Groups of three rats were used for each
sample. The livers were excised immediately after decapi-
tation and pooled for cell fractionation or RNA extrac-
tion. All procedures were carried out in accordance with
European regulations concerning animal experimentation.
Plasmids
Several different fragments derived from the 5¢flanking
region of the CYP3A1 gene (GenBank accession no.
X62086) [28], were previously subcloned upstream of the
thymidine kinase gene [29] in the luciferase expression vector
pT81Luc [30], and analysed in transient transfection
experiments. Constructs bearing multiple element copies
were made using oligonucleotides encompassing the two
distinct CYP3A1 5¢sites 3A1-300 ()350/)331, 5¢-
GTCCTTCTGTAATGGTGTG-3¢), or 3A1-600 ()629/
)608, 5¢-TGCAGGATTGCAGAAGTCTATT-3¢).These
were ligated with the SmaI-digested pT81Luc vector and
analysed in transient transfection experiments. All con-
structs were verified by DNA sequencing. The pMSV/EBPa
and pMSV/EBPbexpression vectors were kindly provided
by S. L. McKnight (University of Texas South-Western
Medical School, TX, USA).
Cell culture
CHO cell line (hamster ovary epithelial) was maintained in
Ham’s F12 medium and the COS-7 cell cultures were
maintained in high glucose Dulbecco’s modified Eagle’s
medium (DMEM) supplemented with 10% (w/v) heat
inactivated fetal bovine serum and maintained at 37 Cin
humidified 5% (v/v) CO
2
.
Transactivation assays
To minimize variations in transfection efficiency, replicates
were transfected in single batch suspension with FuGENE
(Roche Molecular Biochemicals), according to the manu-
facturer’s instructions. Plates containing 150 000 cells were
cotransfected with 0.5 lg of CYP3A1T81Luc plasmid and
different amounts of each expression vector. Cells were
inoculated in 24-well plates and maintained for 48 h. These
cells were harvested and lysed in reporter lysis buffer
(Promega, Madison, WI, USA). Cell extracts were assayed
for luciferase activity and protein content (BCA reagent,
Pierce, Rockfort, IL, USA). The cell extracts were normal-
ized for the total amount of protein prior to performing the
luciferase assay, as C/EBP expression vector was shown to
severely repress the expression of b-galactosidase in the
transfected CHO cell line.
Site-directed mutagenesis
The plasmid 0.8T81Luc served as the template for site-
directed mutagenesis using the QuickChange Site Directed
Mutagenesis Kit (Stratagene, CA, USA). All reactions were
performed according to the protocol provided by the
manufacturer. The oligonucleotides used for site-directed
mutagenesis of the C/EBP binding sites were as follows
(mutated bases underlined): site 3A1-300: 5¢-GGAGA
AAGTCCGTCTATGGTGGTGTGCAGATGACACAG
TTTTGGC-3¢; site 3A1-600: 5¢-GCCTCTGCTCTGTA
AGTGCAGGACCGTAGAGGTCTATTACTTATG-3¢.
mRNA analysis
Total liver RNA was extracted by a modification of the LiCl/
urea method [31], and samples (20 lg) eletrophoresed on
formaldehyde-agarose gels for Northern blot analysis. The
subsequently generated Northern blot nylon membranes
were incubated with the specific oligonucleotide probes
FEBS 2003 Two C/EBP sites in the CYP3A1 locus (Eur. J. Biochem. 270) 557
CYP3A1 (5¢-TGTGCGGGTCCCAAATCCGT-3¢) and
C/EBPa(5¢-GCACGAGACGTCTATAGACA-3¢) end-
labelled with [c-
32
P] dATP using T4 polynucleotide kinase.
Nuclear extracts
Isolation of liver nuclei both from control and dexameth-
asone-treated rats was carried out [32] and nuclear extracts
were prepared as described previously [33].
Recombinant C/EBPaused in the gel mobility-shift assays
were obtained by transfecting 3.0 ·10
6
COS-7 cells with
30 lgofpMSV/EBPa. Cells were seeded in 100-mm plates
and maintained for 48 h for nuclear extract preparation [33].
Western blot analysis
Five micrograms of liver nuclear protein were electrophore-
sed on 10% SDS/polyacrylamide gels and electroblotted onto
Imobilon P (Millipore, Bedford, MA, USA). After visua-
lization of the transferred proteins by amido black staining,
the membranes were incubated with an anti-C/EBPaIg
(14AA, Santa Cruz Biotechnology). Results were quantified
using the
EAGLE
-
EYE
software package (Stratagene).
Electrophoretic mobility-shift assay (EMSA)
Double-stranded DNA probes and competitors were gen-
erated by annealing from the following complementary
single-stranded oligonucleotides: C/EBP consensus oligo-
nucleotide (C/EBPcons), 5¢-TGCAGATTCCGCAATCTG
CA-3¢[34]; 3A1–300, 5¢-AGTCCTTCTGTAATGGTG
TG-3¢; 3A1–600, 5¢-TGCAGGATTGCAGAAGTCTA
TT-3¢; 3A1-700, 5¢-AATTTTGGTGGATAGATAT
AG-3¢; m3A1-300, 5¢-AGTCCGTCTATGGTGGTGTG-3¢;
m3 A1-600, 5¢-TGCAGGACCGTAGAGGTCTATT-3¢
(mutated bases are underlined). The oligonucleotides 3A1-
300 (position )350 to )331), 3A1-600 (position )629 to
)608) and 3A1-700 (position )766 to )746) encompass
distinct CYP3A1 promoter regions. DNA (6 pmol) was end-
labelled with [c-
32
P] dATP using T4 polynucleotide kinase,
and unincorporated nucleotides were removed by Sephadex
G50 filtration. The binding reactions were performed in a
total volume of 20 lL and contained 2–5 lg of nuclear
extract, 10 m
M
Hepes buffer, pH 8.0, 0.1 m
M
EDTA, 2 m
M
dithiothreitol, 17.5% (v/v) glycerol, 40 m
M
spermidine,
40 m
M
MgCl
2
,0.5lgofdIdC,1lg salmon sperm DNA
and 0.5–2 ng of oligonucleotide probe. In competition assays
excess unlabeled oligonucleotide was preincubated (30 min)
at 4 C, prior to incubation with each probe for additional
20 min. Supershift reactions were performed with 1 lLof
anti-C/EBP (14AA, Santa Cruz Biotechnology) which was
added to the reaction media, that were kept on ice for 30 min
before addition of the probe. ProteinÆDNA complexes were
resolved on 5% (w/v) nondenaturing polyacrylamide gels
(acrylamide/bisacrylamide 29 : 1, v/v) in 0.5·Tris/borate/
ECTA buffer (45 m
M
Tris/borate, 1 m
M
EDTA). The gels
were eletrophoresed for 2.5 h, at 30 mA.
Statistical analysis
Statistical analyses were performed using the Student’s t-test
and the
ANOVA
one-way test with the Tukey HSD posthoc
test for unequal N(Spjotvoll/Stoline test). All analysis were
performed using the StatSoft Inc. (1995)
STATISTICA FOR
WINDOWS
software.
Results
Identification of C/EBP-responsive sequences
in the CYP3A1 gene promoter
CYP3A1T81Luc recombinants containing the 5¢upstream
region of the CYP3A1 gene were constructed and cotrans-
fected into CHO cells together with the C/EBP expression
vectors pMSV/EBPaor pMSV/EBPb.TheCHOcellline,
which is devoid of hepato-specific transcription factors, has
been used previously to characterize C/EBP-dependent gene
expression [26,35]. We found that C/EBPasignificantly
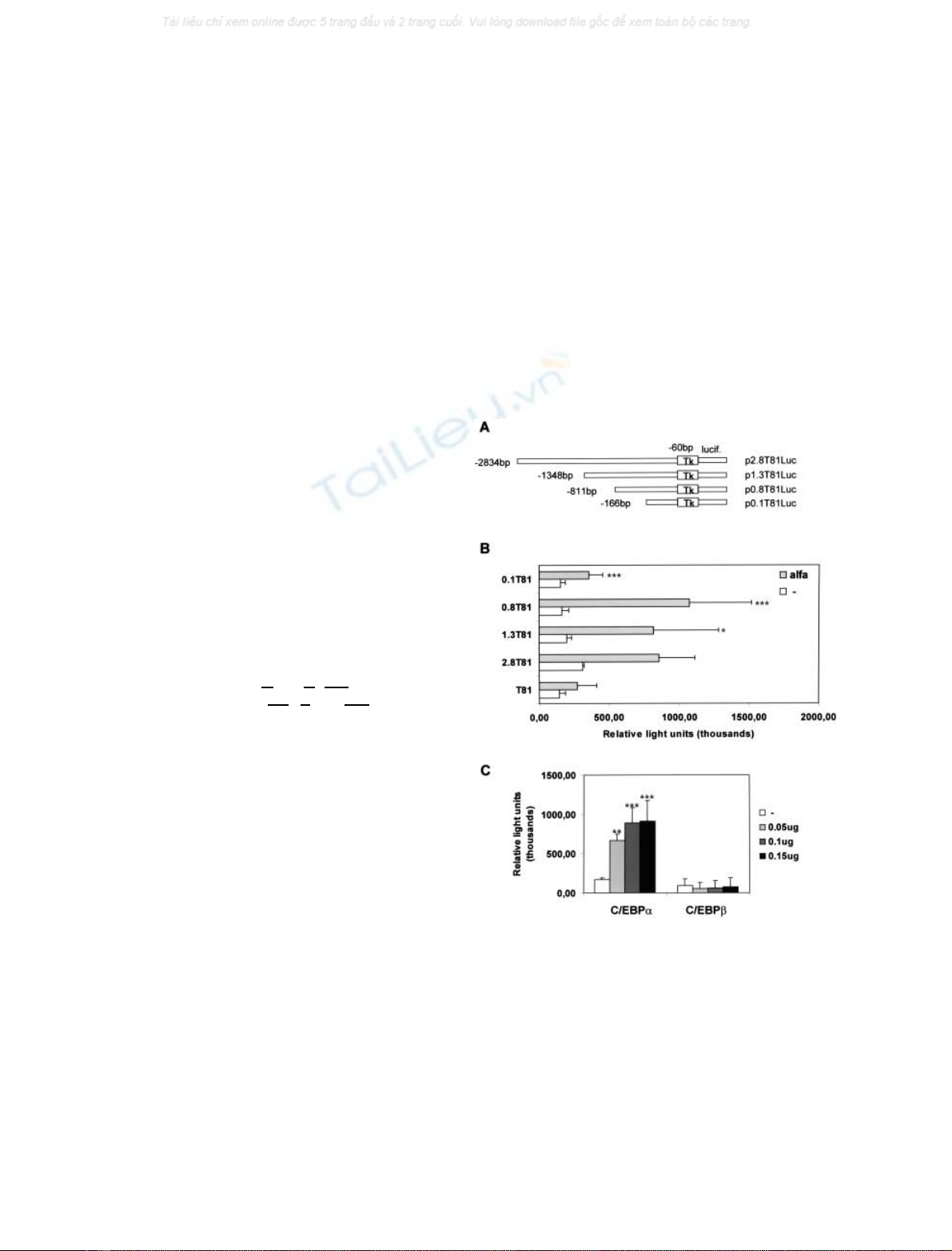
Fig. 1. Transactivation of CYP3AT81Luc constructs with vectors
pMSV/EBPaor pMSV/EBPbin the CHO cell line. Cotransfections
were carried out using 0.5 lg of CYP3AT81Luc and increasing con-
centrations (0.025–0.2 lg) of C/EBP expression vectors, or empty
vector. (A) Diagramatic representation of the luciferase constructs
containing CYP3A1 5¢-flanking DNA sequences, upstream of luci-
ferase cDNA. Tk, thymidine kinase promoter; lucif, luciferase cDNA.
(B) Cotransfection of CYP3AT81Luc with 0.15 lgofpMSV/EBPa.
(C) Cotransfection of p0.8T81Luc with increasing concentrationsof
pMSV/EBPaor pMSV/EBPb. The normalized luciferase activities are
expressed as mean values ± SD of duplicates for a minimum of three
experiments. *P< 0.05, **P<0.01,***P< 0.001 significant dif-
ferences between cells cotransfected with pMSV and pMSV/EBPa
(Student’s t-test).
558 E. Rodrigues et al.(Eur. J. Biochem. 270)FEBS 2003
increases the luciferase reporter gene activity of the
CYP3A1T81Luc (Fig. 1B). The strongest transactivation
level (approximately sevenfold) was observed in the pre-
sence of the p0.8T81-Luc construct (Student’s t-test,
P< 0.001), which contains approximately 800 bp of the
5¢upstream region of the CYP3A1 locus (Fig. 1B). Deletion
of the DNA segment )166 to )811 severely reduced
transactivation by C/EBPa. Moreover, transactivation of
the p0.8T81Luc is shown to be dose-dependent with the
C/EBPaexpression vector (Fig. 1C).
C/EBP-binding activity in rat liver has been described as
involving both C/EBPaand C/EBPb. However, when
pMSV/EBPbwas assayed in cotransfection experiments
with the CYP3A1 recombinant p0.8T81Luc no transacti-
vation was observed (Fig. 1C). Moreover, C/EBPbinhibits
the C/EBPa-stimulated expression of the p0.8T81Luc (data
not shown).
Characterization of the C/EBP binding sequences
in the CYP3A1 promoter region
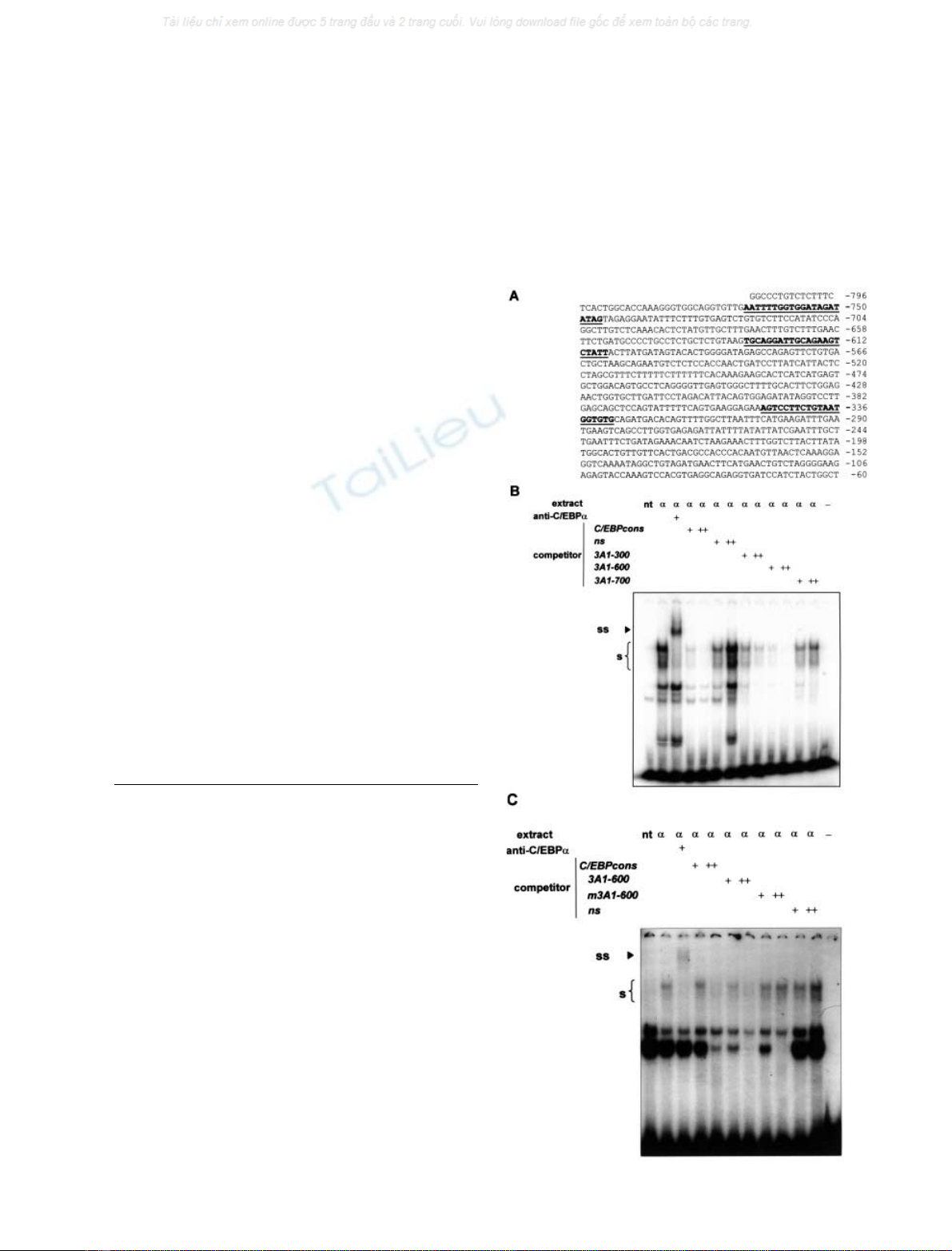
We used gel mobility-shift assays, with the goal of identi-
fying active C/EBPabinding sites within the CYP3A1 locus.
First, oligonucleotides encompassing the putative C/EBP
responsive elements in p0.8T81Luc were synthesized. These
were named 3A1-300, 3A1-600 and 3A1-700 (Fig. 2A).
C/EBP proteins were then over-produced in COS-7 cells,
and the nuclear extracts used to characterize their binding
activities to 3A1-300, 3A1-600 or 3A1-700 by EMSA. The
specificity of the complex(es) formed when a C/EBP
consensus oligonucleotide (C/EBPcons) was used, was
verified by means of a supershift assay using an anti-C/
EBPaIg (Fig. 2B; arrowhead). Both 3A1-300 and 3A1-600
oligonucleotides competed efficiently for C/EBPabinding
to the C/EBPcons preventing the formation of the complex
at a 50-fold molar excess (Fig. 2B). The C/EBPconsÆ
C/EBPacomplex was not competed by 3A1-700 or by an
unrelated DNA sequence (Fig. 2B).
Supershift assays, with an anti-C/EBPaIg and radio-
labelled oligonucleotides 3A1-600 and 3A1-300 (data not
shown) confirmed the specificity of C/EBPaprotein binding
(Fig. 2C; arrowhead). We note that the 3A1-600ÆC/EBPa
complex was competed by C/EBPcons and by cold self,
while a mutant form of this oligonucleotide, differing in
Fig. 2. Characterization of the binding activities to sites 3A1-300, 3A1-
600 and 3A1-700 in C/EBPaover-expressed COS-7 cell nuclear
extracts. (A) DNA sequence of the CYP3A1 5¢-flanking region cor-
responding to construct p0.8T81Luc. The oligonucleotides containing
the putative C/EBP responsive elements used in the gel mobility-shift
assays are underlined. The oligonucleotide 3A1-300 corresponds to the
)350/)331 sequence of the CYP3A1 cDNA, the 3A1-600 to the )629/
)608 sequence and the 3A1-700 to nucleotides )767/)747. (B) EMSA
was performed using a radiolabeled double-stranded oligonucleotide
corresponding to the C/EBP consensus (C/EBPcons) as a probe.
Competition experiments were performed by adding a five- or 50-fold
excess of unlabelled double-stranded oligonucleotides corresponding
to cold-self, a nonspecific tubulin sequence, and 3A1-300, 3A1-600 or
3A1-700. (C) EMSA was performed using a radiolabeled double-
stranded oligonucleotide corresponding to site 3A1-600 as a probe.
Competition experiments were performed by adding a five- or 50-fold
excess of unlabelled double-stranded oligonucleotides corresponding
to C/EBPcons, cold self, mutant 3A1-600 (m3A1-600), and a non-
specific tubulin sequence. Supershift experiments were performed using
an anti-C/EBPaIg. Symbol sin panel B and C denotes the position of
the C/EBP containing complex and the arrowhead ssthe position of
the DNAÆprotein complex shifted by the specific antibody.
FEBS 2003 Two C/EBP sites in the CYP3A1 locus (Eur. J. Biochem. 270) 559
3 bp, did not. The results suggest the presence of two
C/EBPabinding sites at positions )300 and )600 within the
CYP3A1 locus.
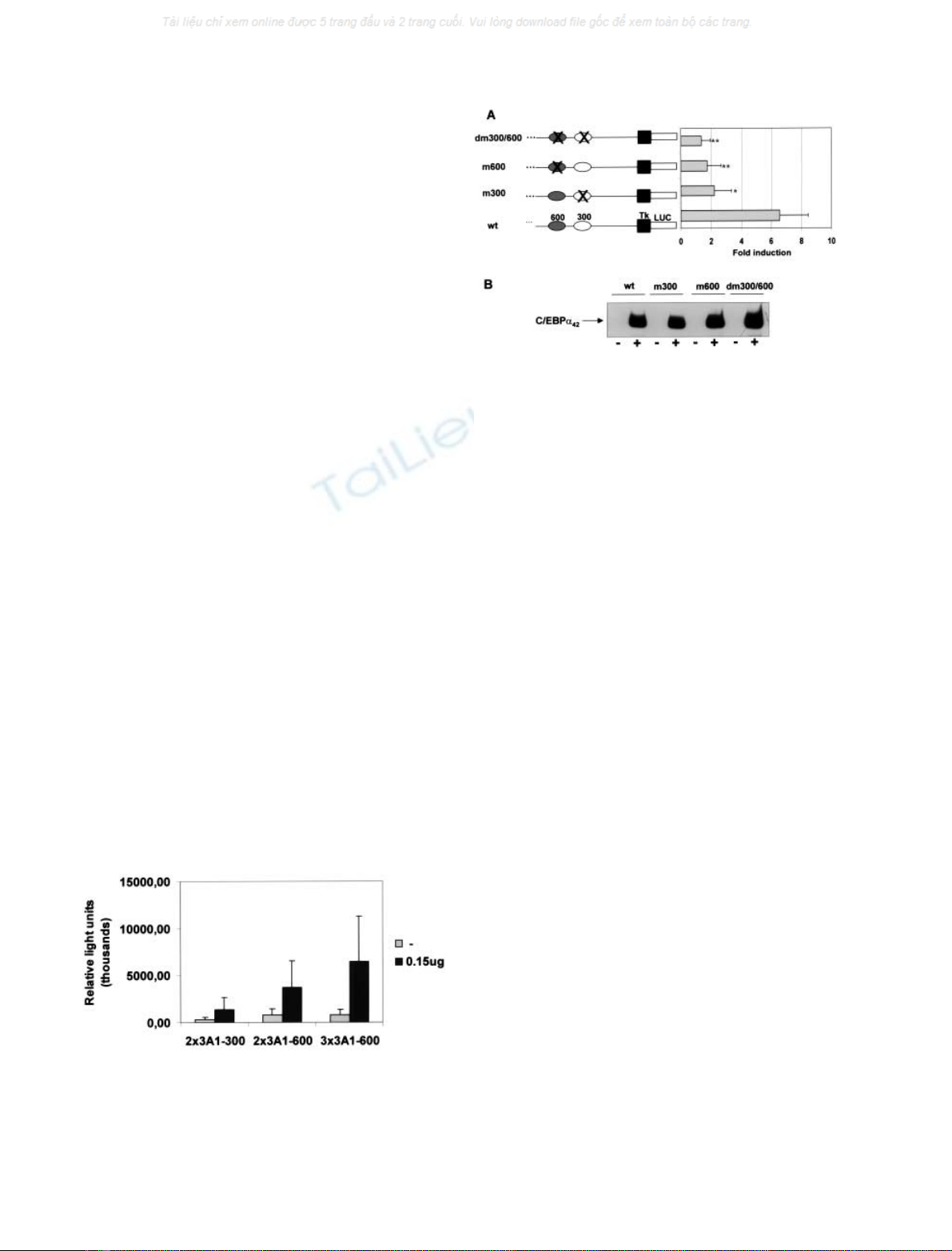
Activation of multimerized CYP3A1-C/EBP responsive
elements by C/EBPain transfection assays
To determine whether C/EBPabinding to either site 3A1-
300 or 3A1-600 resulted in transcriptional activation, we
analysed the ability of each specific site to confer C/EBP
responsiveness to the thymidine kinase promoter of the
pT81Luc recombinant. We performed the cotransfection of
CHO cells with pMSV/EBPaand single recombinants
containing two or three synthetic copies of 3A1-300 or 3A1-
600 oligonucleotides. As shown in Fig. 3, the C/EBP
elements found in the CYP3A promoter are functional
and autonomous units that confer C/EBP responsiveness to
a heterologous promoter.
Functional analysis of C/EBP binding sites in CYP3A1
promoter by site directed mutagenesis
The functional relevance of sites 3A1-300 and 3A1-600 for
CYP3A1 induction by C/EBPawas investigated by
individual or combined mutagenesis of the two sites in
p0.8T81Luc, followed by cotransfection of C/EBPa
expression vector into CHO cells. As shown in Fig. 4,
mutation of site 3A1-300 (m300) and/or 3A1-600 (m600)
significantly reduced activation of the p0.8T81Luc by
C/EBPato approximately 50% of the wild-type level (wt)
(
ANOVA
one-way test: F ¼12.29, d.f. ¼9, P< 0001).
The posthoc comparisons revealed that significant differ-
ences were found between the wt and each mutant (Tukey
HSD for unequal N: m300, P< 0.05; m600, P<0.01;
dm300/600, P< 0.01). Western blot analysis of the
42 kDa C/EBPaprotein in the cotransfection experiments
proved that the decrease in CYP3A1 activation is not due
to a reduction in C/EBPaexpression (Fig. 4B). Taken
together, these results strongly indicate that both sites 3A1-
300 and 3A1-600 are functional elements of the CYP3A1
promoter.
Time-course analysis of the
in vivo
expression
of C/EBPaupon glucocorticoid administration
The time-course variation of the relative concentrations of
C/EBPamRNAandproteinwasmonitoredintheratliver.
In parallel we also followed the accumulation of the
CYP3A1 mRNA, upon addition of dexamethasone
(Fig. 5).
C/EBPamRNA concentration increases approximately
fivefold over the control value between 0.5 and 4 h after
treatment, and decreases to about fourfold of the control
value by 21 h after dexamethasone administration. Such an
increase clearly precedes the marked induction of hepatic
CYP3A1 mRNA found to occur between 4 and 21 h after
treatment (Fig. 5A).
We found the accumulation of both isoforms of C/EBPa
(p42, of 42 kDa and p30, of 30 kDa) to occur concomit-
antly with the increase in the relative abundance of the
corresponding mRNA. However, the two isoforms accu-
mulate with different kinetic profiles (Fig. 5B). Maximal
induction (an increase of about 2.5-fold over the basal level)
is observed for p30 between 2 and 4 h upon dexamethasone
administration, with a gradual decrease to the control value
21 h after treatment. In contrast the levels of the p42
isoform increases from hour 2 onwards, reaching constant
Fig. 3. Ability of multimerized CYP3A1-C/EBP responsive elements to
confer activation by C/EBPain the CHO cell line. Cotransfections were
carried out with 0.5 lgof3A1-C/EBP-RE-T81Luc and 0.15 lgof
pMSV/EBPaexpression vector, or empty vector. The normalized
luciferase activities are expressed as mean values ± SD of duplicates
for a minimum of three experiments.
Fig. 4. Functional analysis of the C/EBP binding sites in the CYP3A1
promoter by cotransfection studies using the p0.8T81Luc reporter plas-
mid altered by site-directed mutagenesis. (A) Cotransfections were
carried out using 0.5 lg of wild-type (wt) or the different mutated
promoter reporter plasmids (mut300, mut600 and dm300/600) and
0.15 lg of pMSV/EBPaexpression vector, or empty vector. As no
significant differences in the basal activity of the different mutated
reporter plasmids were found, normalize luciferase activities are
expressed as mean values ± S.D. of fold induction of duplicates for a
minimum of three experiments. *P< 0.05, **P< 0.01 significant
differences in transactivation due to mutation of 3A1-300 or/and 3A1-
600 sites (
ANOVA
one-way test: F ¼12.29, d.f. ¼9, p < 0001; posthoc
Tukey HSD test for unequal N: m300, P< 0.05; m600, P<0.01;
dm300/600, P< 0.01). (B) Result of a representative Western blot
analysis of the 42 kDa C/EBPaprotein in the cotransfection experi-
ments: lanes (–) with empty vector and (+) with 0.15 lgofpMSV/
EBPa. Fifty lL of total protein extract were analysed by SDS/PAGE
(10%, w/v), the resolved proteins transferred to poly(vinylidene
difluoride) membranes, and the membranes incubated with a specific
anti-C/EBPaIg.
560 E. Rodrigues et al.(Eur. J. Biochem. 270)FEBS 2003


























