
Molecular characterization of
MRG19
of
Saccharomyces cerevisiae
Implication in the regulation of galactose and nonfermentable carbon source
utilization
Firdous A. Khanday*, Maitreyi Saha and Paike Jayadeva Bhat
Laboratory of Molecular Genetics, Biotechnology Center, Indian Institute of Technology, Powai, Mumbai, India
We have reported previously that multiple copies of MRG19
suppress GAL genes in a wild-type but not in a gal80 strain of
Saccharomyces cerevisiae. In this report we show that dis-
ruption of MRG19 leads to a decrease in GAL induction
when S. cerevisiae is induced with 0.02% but not with 2.0%
galactose. Disruption of MRG19 in a gal3 background (this
strain shows long-term adaptation phenotype) further delays
the GAL induction, supporting the notion that its function is
important only under low inducing signals. As a corollary,
disruption of MRG19 in a gal80 strain did not decrease the
constitutive expression of GAL genes. These results suggest
that MRG19 has aroleinGAL regulation only when the
induction signal is weak. Unlike the effect on GAL gene
expression, disruption of MRG19 leads to de-repression of
CYC1-driven b-galactosidase activity. MRG19 disruptant
also showed a twofold increase in the rate of oxygen uptake
as compared with the wild-type strain. ADH2,CTA1,
DLD1,andCYC7 promoters that are active during non-
fermentative growth did not show any de-repression of
b-galactosidase activity in the MRG19 disruptant. Western
blot analysis indicated that MRG19 is a glucose repressible
gene and is expressed in galactose and glycerol plus lactate.
Experiments using green fluorescent protein fusion con-
structs indicate that Mrg19p is localized in the nucleus
consistent with the presence of a consensus nuclear locali-
zation signal sequence. Based on the above results, we pro-
pose that Mrg19p is a regulator of galactose and
nonfermentable carbon utilization.
Keywords: carbon metabolism; CYC1 repressor; GAL genes;
glucose repression; induction signal; transcriptional
regulator.
The reprogramming of molecular machinery mainly
brought about by transcriptional regulation, co-ordinates
different cellular processes as cells move from one physio-
logical state to another. Since this is the key for the
evolutionary success of any organism, it is not surprising
that significant fraction of their genetic endowment is
dedicated to regulatory functions. When yeast shifts from
the most preferred carbon source glucose to galactose, a
large increase in the synthesis of GAL gene products occurs,
without affecting its fermentative life style [1–4]. Obviously,
during this transition, yeast has to make compensatory
changes in the pattern of gene expression to co-ordinate
galactose metabolism with various other cellular processes,
especially energy metabolism. One of the obvious changes is
the de-repression of many glucose-repressed functions,
especially mitochondrial biogenesis [5–8]. Recently,
genome-wide analysis has identified genes which previously
were not suspected to be induced in presence of galactose,
emphasizing the importance of the need for multiple
pathways to integrate various cellular functions [9]. Study
of utilization of galactose by Saccharomyces cerevisiae
provides a convenient experimental system to probe into the
network of gene interaction leading to exquisite co-ordina-
tion between different cellular processes [10].
Gal4p, a DNA binding transcriptional activator, acti-
vates the GAL genes in response to galactose. Although
Gal4p remains bound to the upstream activating sequences
of GAL genes in noninducing conditions, Gal80p inhibits
transcriptional activation. This is due to a physical interac-
tion between Gal4p and Gal80p [11]. In response to
galactose, Gal3p interacts with Gal80p, thereby allowing
Gal4p to cause rapid transcription of GAL genes
[1,2,4,12,13]. The long-term adaptation phenotype exhibited
by a gal3 strain [14], is due to Gal1p, which has Gal3p-like
signal transduction activity in addition to galactokinase
activity [15]. Recent experiments have demonstrated that
Gal3p directly interacts with Gal80p in the presence of
galactose and ATP [16–19]. It has also been demonstrated
that a tripartite complex is formed between Gal3p-Gal80p-
Gal4p in response to galactose and ATP [3]. The current
view is that the interaction of Gal3p with Gal80p allows the
transcription-activating domain of Gal4p to interact with
the general transcription factors, thereby causing transcrip-
tion activation of GAL genes [20,21]. It has been suggested
that the interaction of Gal3p with Gal80p may not result in
the dissociation of Gal80p from Gal4p [22] but may cause
Gal80p to shift to a second site on Gal4p [19]. Based on the
results that Gal3p is cytoplasmic and Gal80p is distributed
in both the nucleus and the cytoplasm, it has been suggested
Correspondence to P. J. Bhat, Laboratory of Molecular Genetics,
Biotechnology Center, Indian Institute of Technology, Powai,
Mumbai 400 076, India.
Fax: + 91 22 572 3480, Tel.: + 91 22 576 7772,
E-mail: jayadeva@btc.iitb.ac.in
Abbreviation: IPTG, isopropyl thio-b-
D
-galactoside; YEP, yeast
extract peptone.
*Present address: School of Medicine, John Hopkins University,
Baltimore, MD 21205, USA.
(Received 15 July 2002, revised 27 September 2002,
accepted 10 October 2002)
Eur. J. Biochem. 269, 5840–5850 (2002) FEBS 2002 doi:10.1046/j.1432-1033.2002.03303.x

that the dynamics of their distribution is intrinsic to GAL
gene regulation [23]. Recent studies have indicated that the
Gal80p–Gal80p interaction is required for complete repres-
sion of GAL genes [24].
Circumstantial evidence suggests that the energy status of
the cell is an important determinant of GAL gene induction
[15,25–27]. This suggests that the availability of metabolic
energy is a rate-limiting step in the synthesis of GAL
enzymes that constitute 5% of the total soluble proteins
when the cell grows on galactose as sole carbon source
[28,29]. Phosphorylation of S699 of Gal4p has been shown
to be important for activating transcription of GAL genes
when the induction signal is weak and has been suggested to
be a link between energy status and the GAL genetic switch
[27]. Although, the importance of mitochondrial function in
galactose metabolism has been well recognized, the
molecular basis for the same has largely remained unex-
plored. We had reported the isolation of MRG19 as a
multicopy suppressor of galactose toxicity at low but not at
high induction signal [30]. Results presented in this
communication indicate that Mrg19p is a regulator of
GAL and CYC1 expression. We present evidence that
Mrg19p is an integral component required for the maximal
induction of GAL when the induction signal is weak.
Results indicate that Mrg19p is a canonical repressor of
CYC1. Based on the above, we propose that Mrg19p
regulates fermentation and aerobic oxidation.
MATERIALS AND METHODS
Strains, media and growth conditions
Table 1 provides the details of yeast strains used in this
study. Yeast strains were grown at 30 Cinrichyeast
extract peptone (YEP) or defined synthetic drop-out or
synthetic complete media as described [31]. Carbon sources
were added to YEP, synthetic drop-out or synthetic
complete media to a final concentration of 2% w/v glucose,
2% or 0.02% galactose and/or 3% glycerol plus 2%
potassium lactate (v/v) pH 5.7. Yeast transformations were
carried out as described [32]. Escherichia coli strain XL1-
Blue was used for plasmid construction and amplification.
Bacterial transformation was carried out as described [33].
E. coli strain BL21 (DE3) was used for expression of fusion
protein from pET32(a). E. coli XL1-Blue and BL21 (DE3)
strains were grown at 37 C in Luria–Bertani broth with
ampicillin at a final concentration of 75 lgÆmL
)1
wherever
required for plasmid maintenance [34]. For the induction of
fusion protein in BL21 (DE3), isopropyl thio-b-
D
-galacto-
side (IPTG) was added to a final concentration of 1 m
M
at
an OD
600
of 0.5 and growth was allowed to continue for a
further 2 h.
Plasmids
A4.7kbHindIII–SalI fragment obtained from YIp24
ADH2-lacZ(+) [35], containing ADH2::lacZ cassette, was
subcloned into HindIII–SalIdigestedYCplac33 [36] and the
resulting plasmid was named as YCfADH2::lacZ. A 4-kb
XbaI fragment was obtained from plasmid pAB2654
(unpublished data) containing the CYC7::lacZ cassette and
was subcloned into XbaIdigestedYCplac33; the resulting
plasmid was named YCfCYC7::lacZ. A 5.6-kb PstI–SalI
fragment obtained from YIpCTA1-lacZ [37], containing the
CTA1::lacZ cassette, was subcloned into PstI–SalIdigested
YCplac33 and the resulting plasmid was named pYCfCTA
1::lacZ. A portion of MRG19 was amplified by PCR using
primers PJB102 (5¢-GACCGTAGGTACCATGTTGGCT
TCAG-3¢) and PJB103 (5¢-CGGGCCCCTC GAGGCCCA
TCATCTAA-3¢) carrying KpnIandXhoI sites, respectively.
After digesting the PCR product with KpnIandXhoI, it was
cloned into KpnI–XhoI digested pET32a and the resulting
plasmid was named p19C-KX. The protein product obtained
from the above construct upon induction with IPTG was
found to be 67 kDa as expected. As the induction of this
truncated protein was low, a frame-shift mutation was
introduced in p19C-KX by digesting with SalI and filling in
with dNTPs and the resulting plasmid was named p19C-S.
This construct was expected to induce a protein of 49 kDa.
To determine subcellular localizations of Mrg19p, two
in-frame fusion constructs with GFP were made. A 2.9-kb
SmaI–SalIfragmentofMRG19 was subcloned into SmaI–
SalI digested pGFP-N-FUS [38] and the resulting plasmid
was named pGFP-N-19FUS. pGFP-N-19FUS was further
digested by SmaI–HindIII to remove the nuclear localization
signal (NLS) and the resulting plasmid was named pGFP-
N-NLSFUS.
Strain constructions
A derivative of ScPJB644 with LEU2 was constructed as
follows. ScPJB644 was transformed to leucine prototrophy
with a 5.4-kb genomic fragment containing LEU2 gene,
which was isolated by digesting YEp13 with PstI. The
Table 1. List of strains.
Name Genotype Source
Sc289-1 MATaura3-52 trp1-289 gal7Dgal1DLaboratory stock
Sc285 MATa ura3-52 leu2-3, 112 gal80 J.E. Hopper
Sc285-19DMATa ura3-52 leu2-3, 112 gal80 mrg19:: LEU2 This study
ScPJB644-L MATa ura3-52 leu2:: LEU2 trp1 This study
ScPJB644-19DMATa ura3-52 leu2-3112 trp1, mrg19::LEU2 Laboratory stock
ScPJB644-19DMATaura3-52 leu2-3, 112 trp1 mrg19::LEU2 This study
Sc385 MATa ura3-52 leu2-3, 112 ade1 ile, MEL1 GAL3::LEU2 J.E.Hopper
Sc385-19DMATa ura3-52 leu2-3, 112 ade1 ile MEL1, GAL3::LEU2, mrg19::LEU2 This study
H190 MATa SUC2 ade2-1 can1-100 his3–11,15, leu2-3112 trp1-1 ura3-1 mig1-€
aa2::LEU2 H. Ronne
W303-1DMATaSUC2 ade2-1 can1-100 his3-11,15, leu2-3112 trp1-1 ura3-1 H. Ronne
FEBS 2002 MRG19 as a bi-functional regulator (Eur. J. Biochem. 269) 5841

mating type of ScPJB644–19DMATa was changed to
MATaby transforming with HO plasmid as described [15].
Sc285–19Dwas constructed by crossing Sc285 with
ScPJB644–19Dof the opposite mating type.The diploids
selected on synthetic complete glucose medium lacking
leucine and tryptophan were then sporulated [39]. After
digesting the asci with cell wall degrading enzyme, random
spores were screened on synthetic complete glycerol plus
lactate medium lacking leucine to identify disrupted
MRG19 locus, but containing 2-deoxygalactose to identify
gal80 allele [40]. Sc385–19Dwas constructed by mating
Sc385 with ScPJB644–19Dof the opposite mating type.The
diploids selected in synthetic complete galactose medium
lacking tryptophan were sporulated. Leu+ segregants
which are gal3mrg19 were isolated from tetrads of the
constitution 2
+
:2
–
::LEU
+
:LEU
–
by tetrad dissection.
Expression of truncated Mrg19p and generation of
polyclonal antibodies
Antibodies against Mrg19p were raised as described [15].
The cells obtained after induction were treated with SDS gel
loading buffer, boiled for 5 min and subjected to analytical
SDS/PAGE. E. coli strain bearing parent vector (pET32a)
or the plasmid construct (p19C-S) with and without IPTG,
respectively, served as the controls. As expected, a protein of
molecular weight 49 kDa was induced from transformant
bearing the p19C-S in the presence, but not in the absence,
of IPTG. For immunization, a protein of molecular mass
49 kDa was isolated using preparative SDS/PAGE fol-
lowed by electro-elution and then precipitated by acetone.
After collecting blood (to obtain preimmune serum), 100 lg
protein along with Freund’s complete adjuvant was injected
subcutaneously at more than one spot into albino rabbits.
Two weeks after the primary injection, three booster doses
of 100 lg protein were given in incomplete Freund’s
adjuvant. One week after the last booster dose, rabbit was
bled through the marginal vein. Serum was collected after
allowing clot formation at room temperature for 1 h
followed by centrifugation.
Western blot analyses
Cells were harvested by centrifuging at 5000 gfor 5 min and
washed once with cold autoclaved double distilled water.
Whole cell extracts were prepared in the presence of
protease inhibitor cocktail and phenylmethanesulfonyl
fluoride as described. Supernatant obtained from the whole
cell extract was treated with polyethyleneimine to a final
concentration of 0.03% and then centrifuged at 4 Cat
10 000 gfor 2 min. Protein was estimated as described [41].
Supernatant obtained from the above step was kept in a
boiling water bath with gel loading buffer for 5 min and was
subjected to SDS/PAGE on a 7.5% gel. An equal amount
of protein was loaded in all lanes. Proteins were transferred
onto nitrocellulose membrane and blocked with buffer
containing 1% milk powder for 1 h. The blot was then
probed with 1 : 2000 diluted antiserum or preimmune
serum and incubated for 1 h. Membrane was washed with
buffer four times for 5 min each. The immunoblot was
probed with 1 : 2500 diluted secondary antibody conjugat-
ed with alkaline phosphatase. All the experiments were
repeated at least three times.
Galactokinase assay
Cells were washed and extracts were prepared by the glass
bead cell disruption method [4]. Galactokinase activity was
assayed as described [28].
14
C-galactose (58 mCiÆmmol
)1
)
was from Amersham. The original stock of
14
C-galactose
was diluted with cold galactose to achieve a final specific
activity of 1 lCi per 4.7 lmol. DE81 ion exchange paper
was from Whatman International Ltd. The radioactivity
was counted in an LKB liquid scintillation counter using
OCS liquid scintillant (Amersham). Each value is an
average of four independent colonies and the assays were
performed in triplicate.
b-Galactosidase assay
b-Galactosidase activity was assayed in cell extracts as
described [39]. Duplicate samples were taken for each
determination. Experiments were performed with five
independent transformants and the result of four different
experiments is presented. Protein was estimated by the
Bradford method. Specific activities are represented as nmol
product formedÆmin
)1
Æmg protein
)1
.
Analysis of O
2
consumption
Cells grown on glycerol plus lactate as carbon source were
harvested either in the log phase or in the stationary phase.
The cells were washed three times with ice-cold distilled
water; the wet weight of the pellets was determined and
resuspended in oxygraph buffer [1% yeast extract, 0.1%
K
2
HPO
4
,0.12%(NH
4
)
2
SO
4
(pH 4.5)] at 100 mg cellÆmL
)1
.
Oxygen consumption rates were measured using a Clark-
type oxygen electrode, with 0.1 m
M
ethanol as substrate.
The rates were measured from the slope of a plot of O
2
concentrationvs.timeandexpressedasnmolO
2
consumed
per min per 10 mg wet weight of cells [42].
GFP fluorescence microscopy
Wild-type cells were transformed with pGFP-N-19FUS and
pGFP-N-NLSFUS plasmids. Cells were grown to D
600
of
1.5 in methionine and uracil double drop-out glycerol (3%)
lactate (2%) media [38]. Cells were allowed to grow with
4¢,6-diamidino-2-phenylindole (DAPI) at a concentration
2lgÆmL
)1
for 1 h before microscopic observation. Cells
were harvested in the cold and green fluorescent protein
(GFP)/DAPI fluorescence was monitored using a Zeiss
LSM510 Scanning Confocal Microscope. Images were
recorded and processed in
ADOBE PHOTOSHOP
6.0.
RESULTS
MRG19
as a regulator of galactose utilization
MRG19 disruptant is defective in galactokinase expres-
sion in response to galactose. Recently, it was shown that
the activity of wild-type GAL4 is not different whether 0.02%
or 2.0% galactose is used for induction. However,
GAL4S699 A is defective in GAL gene induction at 0.02%
but not 2% galactose, indicating a difference in the galactose
signalling mechanism [27]. As MRG19 was isolated as a
multicopy suppressor of galactose toxicity at low galactose
5842 F. A. Khanday et al. (Eur. J. Biochem. 269)FEBS 2002
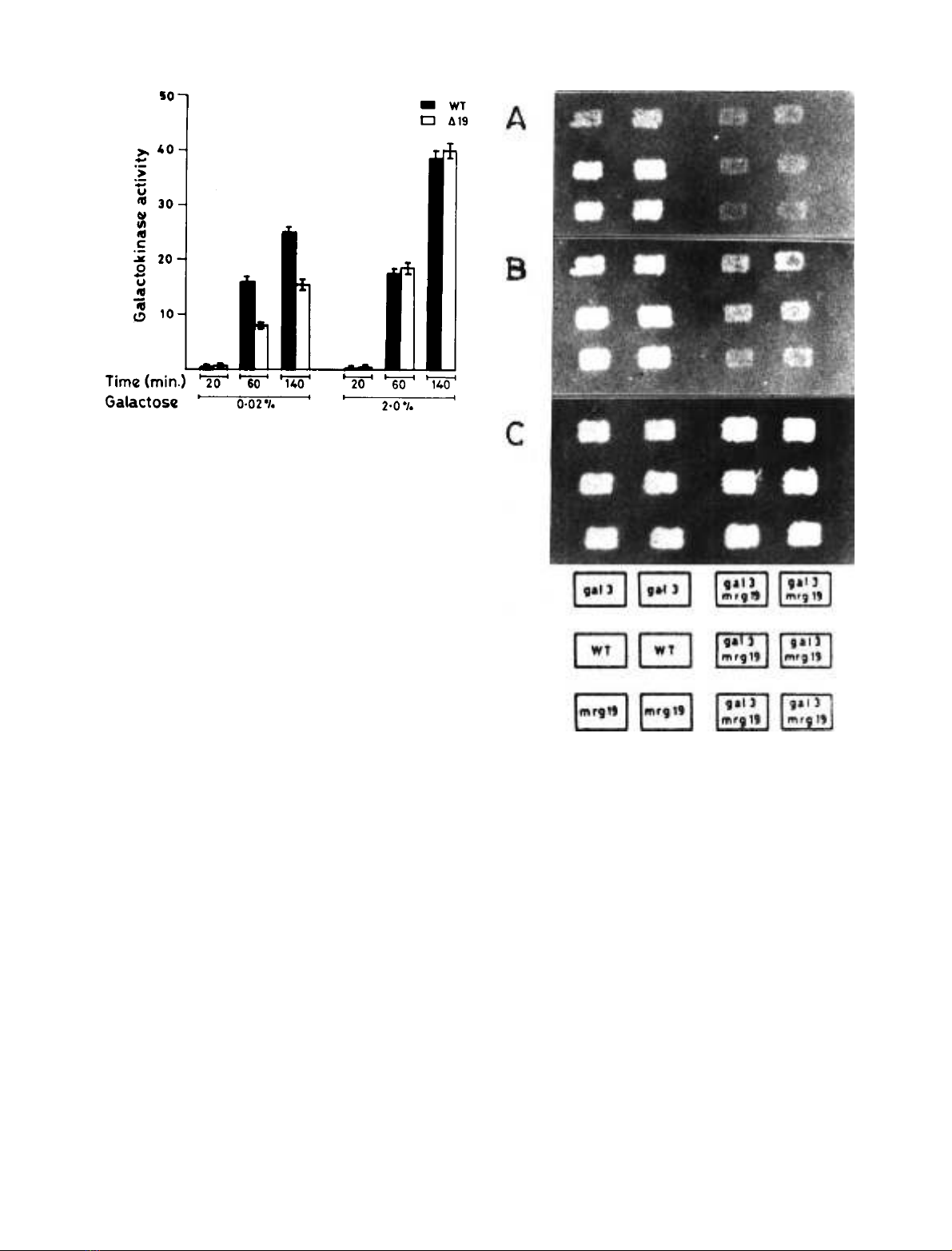
concentration [30], we surmised that disruption of MRG19
might effect GAL induction only at low galactose concen-
trations. This hypothesis was tested by monitoring galac-
tokinase induction as a function of time in the wild-type and
in the MRG19 disruptant when cells were induced by either
0.02% or 2.0% galactose. It is clear from the results that
galactokinase activity is reduced by 50% in the disruptant
as compared with the wild-type, only when the cells were
induced by 0.02% galactose (Fig. 1). These results indicate
that MRG19 is required for maximal GAL gene induction
under conditions when the induction signal is weak.
Disruption of MRG19 in a gal3 background leads to a
delay in long-term adaptation phenotype. To further
investigate the idea that MRG19 function is necessary for
maximum expression of GAL genes only under conditions
when the induction signal is weak, GAL gene expression was
monitored in both a gal3 and a gal3mrg19 strain. The
delayed induction of GAL genes in a gal3 strain is due to the
weak induction signal transduced by the GAL1 gene [15].
Therefore, it was expected that disruption of MRG19 in a
gal3 strain (i.e. gal3mrg19) would not show any change in
the GAL gene expression if the two lie in the same induction
pathway. Alternatively, if they lie in different induction
pathway, gal3mrg19 would exhibit a further delay or may
not induce the GAL gene expression at all. The growth
pattern of wild-type, gal3,mrg19 and gal3mrg19 strains was
monitored as a function of time on complete medium
containing galactose as a carbon source. Wild-type and
mrg19 strains grew on galactose plates within 12 h while
gal3 strain showed the characteristic delay in growth on
galactose. Interestingly, the gal3mrg19 strain showed a
further delay in growth on galactose plate as compared with
the gal3 strain (Fig. 2).
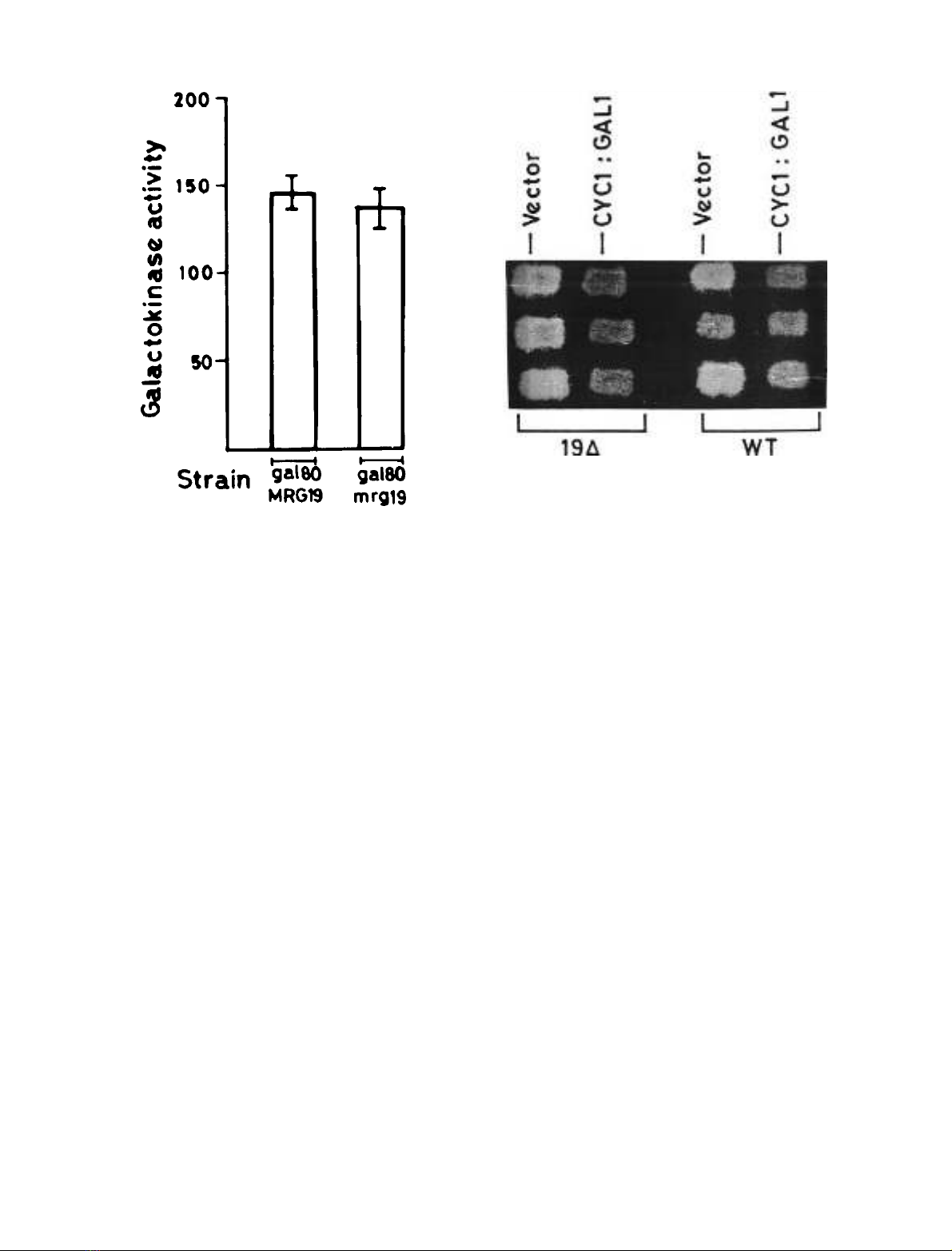
Disruption of MRG19 in a gal80 background does not
affect constitutive GAL gene expression. The results
described above indicated that disruption of MRG19
affects expression of GAL genes only when the induction
signal is weak. This implied that loss of MRG19 function
might not affect the constitutive GAL gene expression
observedinagal80 strain (strong induction signal). To
determine whether this is true or not, galactokinase
activity was determined in gal80MRG19 and gal80mrg19
strain grown in glycerol plus lactate. As expected,
disruption of MRG19 in a gal80 strain (gal80mrg19
strain) did not cause a discernible difference in galacto-
kinase activity (Fig. 3) in comparison with a gal80 strain
(gal80MRG19 strain). This suggested that in a wild-type
strain it is only when the induction signal is weak that
the function of MRG19 is necessary for maximal GAL
gene expression.
Fig. 1. Specific activity of galactokinase in wild-type cells and the
MRG19 disruptant. Cells were grown to D
600
of 0.5 in synthetic me-
dium containing glycerol plus lactate and galactose was added to the
culture to a final concentration of 0.02% or 2.0%. After galactose
addition, cells were allowed to grow for 20, 60 and 140 min Galacto-
kinase activity was determined as described in Materials and methods.
Specific activity is represented as nanomoles of [
14
C]galactose phos-
phorylatedÆmin
)1
Æmg protein
)1
.
Fig. 2. Delayed long-term adaptation phenotype of the mrg19gal3
strain. Wild-type, gal3,mrg19 (in duplicate) and six independent
segregants of genotype gal3mrg19 obtained from three tetrads, were
grown on synthetic complete medium containing 2% glucose and
replica plated onto synthetic complete media containing 2% galactose.
Cells in (A), (B) and (C) were allowed to grow on synthetic complete
media containing 2% galactose for 20, 35 and 50 h, respectively.
FEBS 2002 MRG19 as a bi-functional regulator (Eur. J. Biochem. 269) 5843
MRG19
as a regulator of iso-1-cytochrome C
Disruption of MRG19 results in the de-repression of the
CYC1 promoter. We reported previously that multiple
copies of MRG19 suppress CYC1 driven galactokinase [30].
If MRG19 is a canonical repressor of CYC1,thenitis
expected that disruption of MRG19 would result in the
de-repression of CYC1 promoter. To determine this, we
used a plate assay which is based on the observation that
2-deoxygalactose is toxic to wild-type yeast strains that
constitutively express galactokinase, due to the accumula-
tion of 2-deoxygalactose-1-phosphate [40]. Growth of a
wild-type strain bearing the CYC1::GAL1 construct which
expresses galactokinase at a basal level on glycerol
plus lactate was marginally reduced in the presence of
2-deoxygalactose as compared to its vector control (Fig. 4)
due to 2-deoxygalactose toxicity. If MRG19 disruption
leads to a de-repression of the CYC1 promoter, we would
expect mrg19 transformed with CYC1::GAL1 to show a
diminished growth as compared with the wild-type control.
Growth of an MRG19 disruptant bearing the
CYC1::GAL1 construct was lower than that of the wild-
type transformed with same construct (Fig. 4). Moreover,
growth of the MRG19 disruptant bearing CYC1::GAL1
was lower than that of the vector control. These results
indicated that the disruption of MRG19 de-repressed the
CYC1 promoter.
Genome-wide expression analysis showed that MRG19
transcript levels increased fourfold during diauxic growth
[43], indicating that its function may be important at
higher cell density. Therefore, in the MRG19 disruptant,
one would expect the CYC1 promoter to be de-repressed
to a greater extent at higher cell density than at a lower
cell density. To test the above prediction, CYC1 driven
b-galactosidase activity was monitored in wild-type and
the MRG19 disrupted strain. b-galactosidase activity in
the MRG19 disruptant was twofold higher than that in
the wild-type strain (Fig. 5A) only at a higher cell
density.
To corroborate the above conclusion, we monitored the
rate of oxygen uptake in log and stationary phase cultures
of wild-type and MRG19 disruptant cells. The rate of
oxygen uptake was increased in wild-type and MRG19
disruptant cells in response to exogenously added ethanol
indicating that the cells are able to metabolize the carbon
source (Fig. 6, Compare 1 and 2 or 3 and 4) However, an
increase of 50% in the rate of oxygen uptake was
observed in the MRG19 disruptant as compared with the
wild-type in the absence of exogenously added ethanol
(Fig. 6, compare 1 and 3). A similar pattern was observed
even in the presence of exogenously added ethanol (Fig. 6,
compare 2 and 4). The rate of oxygen uptake in wild-type
and MRG19 disruptant cells obtained from log phase
cultures was indistinguishable either in absence or in the
presence of exogenously added ethanol (data not shown).
The above result is consistent with the observation that
CYC1 is de-repressed in mrg19 disruptant cells only at
stationary phase.
Effect of disruption of MRG19 on b-galactosidase activity
driven by promoters, which are active in a nonfermentable
carbon source. Since disruption of MRG19 de-represses the
CYC1 promoter, we expected that it might also de-repress
promoters that are active in the presence of a nonferment-
Fig. 3. Galactokinase activity in gal80MRG19 and gal80mrg19 strains.
Cells were grown to D
600
of 0.5 in synthetic complete medium con-
taining glycerol plus lactate and galactokinase activity was determined
asdescribedinMaterialsandmethods.
Fig. 4. Expression of galactokinase driven by the CYC1 promoter in the
wild-type strain and the MRG19 disruptant. Transformants (in tripli-
cates) of wild-type and MRG19 disruptant bearing either vector
(control) or CYC1::GAL1 construct were grown in Trp drop-out
synthetic minimal medium containing glucose and were replica plated
on to Trp drop-out synthetic minimal medium containing glycerol plus
lactate supplemented with 0.03% of 2-deoxygalatcose.
5844 F. A. Khanday et al. (Eur. J. Biochem. 269)FEBS 2002


























