Analysis of the effect of potato carboxypeptidase inhibitor
pro-sequence on the folding of the mature protein
Sı
´lvia Bronsoms, Josep Villanueva, Francesc Canals, Enrique Querol and Francesc X. Aviles
Institut de Biotecnologia i Biomedicina and Departament de Bioquı´mica i Biologia Molecular, Universitat Auto
`noma de Barcelona,
Spain
Protein folding can be modulated in vivo by many factors.
While chaperones act as folding catalysts and show broad
substrate specificity, some pro-peptides specifically facilitate
the folding of the mature protein to which they are bound.
Potato carboxypeptidase inhibitor (PCI), a 39-residue pro-
tein carboxypeptidase inhibitor, is synthesized in vivo as a
precursor protein that includes a 27-residue N-terminal and
a seven-residue C-terminal pro-regions. In this work the
disulfide-coupled folding of mature PCI in vitro has been
compared with that of the same protein extended with either
the N-terminal pro-sequence (ProNtPCI) or both N- and
C-terminal pro-sequences (ProPCI), and also with the
N-terminal pro-sequence in trans (ProNt + PCI). No
significant differences can be observed in the folding kinetics
or efficiencies of all these molecules. In addition, in vivo
folding studies in Escherichia coli have been performed using
wild-type PCI and three PCI mutant forms with and without
the N-terminal pro-sequence, the mutations had been pre-
viously reported to affect folding of the PCI mature form.
The extent to which the native-likeform was secreted to the
media by each construction was not affected by the presence
of the N-terminal pro-sequence. These results indicate that
PCI does not depend on the N-terminal pro-sequence for its
folding in both, in vitro and in vivo in E. coli.However,
structural analysis by spectroscopy, hydrogen exchange
and limited proteolysis by mass spectrometry, indicate the
capability of such N-terminal pro-sequence to fold within
the precursor form.
Keywords: pro-region; protein folding; structure; disulfide;
protease inhibitor.
Proteins contain within their amino acid sequence the
required information for their folding. However, other
factors may be required for a fast and efficient folding
in vivo. Molecular chaperones facilitate folding by decreas-
ing the tendency of partially folded proteins to go into non-
productive pathways [1]. The protein disulfide isomerase
and the peptidyl-prolyl-isomerase can also function as
folding catalysts [2,3]. Apart from these components that
have a broad substrate specificity, the folding process may
be also affected specifically by the precursor protein. Many
proteins are synthesized in vivo as precursors in the form of
prepro-proteins. Pre- or signal peptides are often involved
in sorting, while pro-peptides or pro-regions can regulate
many processes [4]. Depending on their function they can be
classified in two groups [5]: the class I pro-peptides, which
are required for the correct folding of the proteins to which
they are attached [6–8] and the class II pro-peptides, which
influence other cellular processes, such as secretion, protein
activity or molecular assembly [9].
Class I pro-peptides have also been termed as intra-
molecular chaperones, and their role in folding has been
demonstrated both in vitro and in vivo [7]. There are not
many examples of the role of the pro-regions in small
disulfide-rich proteins. In these proteins, the folding process
differs from that of larger proteins in that is strongly
constrained by the formation of the disulfide bridges.
Among the most studied proteins of this group we find the
bovine pancreatic trypsin inhibitor (BPTI). Its N-terminal
pro-region contains a cysteine residue which appears to
increase both the yield of properly folded mature BPTI and
therateofthefoldingprocessin vitro [10], providing an
intramolecular thiol-disulfide catalyst. Nevertheless, it did
not appear to have any positive effect under physiological
conditions [11]. Similarly, the pro-region of the guanylyl
cyclase activating peptide (GCAPII) contributes signifi-
cantly to the correct disulfide-coupled folding of the mature
protein and the dimerization of the molecule [12]. In
contrast, the studies performed with x-conotoxins demon-
strated that the mature forms of these molecules contain
sufficient information to direct their folding and correct
disulfide pairing in vitro [13,14]. In all cases, the in vitro
folding of the mature disulfide-rich protein is characterized
by a low efficiency and a slow kinetics. This fact suggests
that these proteins need other factors, apart from their
mature amino acid sequence, in order to fold efficiently and
rapidly into the native form in vivo. The nature of these
factors, whether they are found in their pro-region or in
Correspondence to F. X. Aviles and J. Villanueva, Institut de
Biotecnologia i Biomedicina, Universitat Auto
`noma de Barcelona,
08193 Bellaterra (Barcelona), Spain.
Fax: + 34 93 5812011, Tel.: + 34 93 5811315,
E-mail: fxaviles@einstein.uab.es and villanj1@mskcc.org
Abbreviations: BPTI, bovine pancreatic trypsin inhibitor; CPA, carb-
oxypeptidase A; Cys-Cys, cystine; PCI, potato carboxypeptidase
inhibitor; ProNtPCI, PCI with the N-terminal pro-sequence; ProPCI,
PCI with the N- and C-terminal pro-sequences; ProNt + PCI, PCI
with the N-terminal pro-sequence added in trans; RP-HPLC, reversed-
phase high performance liquid chromatography.
(Received 8 May 2003, revised 7 July 2003,
accepted 16 July 2003)
Eur. J. Biochem. 270, 3641–3650 (2003) FEBS 2003 doi:10.1046/j.1432-1033.2003.03754.x

other cellular components (protein disulfide isomerases,
molecular chaperones …), seems to depend on each
individual protein.
Potato carboxypeptidase inhibitor (PCI) is a 39-residue
globular protein that inhibits several metallocarboxypep-
tidases [15]. It has a 27-residue central core with three
disulfide bridges that forms a T-knot scaffold, also found in
other proteins such as many growth factors [16]. This
molecule is synthesized as a prepro-protein that, besides
the 39-residue mature protein, contains a long 27-residue
N-terminal pro-region of unknown function and a seven-
residue C-terminal pro-region, probably involved in trans-
port to the vacuoles [17]. The folding and unfolding
pathways of mature PCI have been previously studied by
our group and are well characterized [18,19]. The extremely
inefficient folding of PCI in vitro [18], together with the
presence of the above-mentioned long pro-sequences,
suggest a possible involvement of them in the in vivo folding
of PCI.
Here, using different protein variants, we have investi-
gated the role of both pro-sequences in the in vitro refolding
of PCI, together with studies of the influence of the
N-terminal pro-region on its in vivo expression. Both studies
conclude that the pro-regions do not significantly influence
the folding of PCI.
Experimental procedures
Plasmid constructs and mutagenesis
A plasmid containing a synthetic gene encoding for the
major isoform of PCI (IIa)[20] cloned into pINIII-OmpA3
vector [21], was used as a template to generate the plasmidic
constructions used for the expression of the PCI forms
studied. ProNtPCI was obtained by means of one-step
PCR, subcloned into pGEM-T Vector System (Promega),
restricted with XbaIandEcoRI and ligated into pIN-III-
OmpA3 vector. Similarly, this construction was used as a
template to generate the D3, Y37G and G35P/P36G
ProNtPCI mutant genes, by means of one-step PCR.
Constructs for D3, Y37G and G35P/P36G PCI mutant
genes were achieved by PCR of wild-type mature PCI [22].
ProNtPCI was also cloned into pBAT4 expression vector
[23], derived from the pET plasmids [24], with and without
the leader sequence OmpA. ProPCI was generated from
ProNtPCI by means of one-step PCR and cloned into
pBAT4 vector without the leader sequence OmpA. All
constructs cloned into pINIII vector were transformed into
Escherichia coli strain MC1061 and those cloned into
pBAT4 vector were transformed into E. coli strain
BL21(DE3).
Protein expression and purification
For constructs cloned into pINIIIOmpA3 vector, E. coli
MC1061 cells were grown at 37 C and expression was
induced at 0.1 attenuation unit at 550 nm by the addition of
1m
M
isopropyl thio-b-
D
-galactoside, and they were har-
vested by centrifugation (13 000 gat 4 C for 45 min) 20 h
after induction of protein expression. ProNtPCI was
purified from the culture medium by a Sep-Pak C
18
(Waters) cartridge and eluted with 70% isopropanol
containing 0.1% trifluoroacetic acid. The protein was
finally purified by RP-HPLC, on a protein C4
0.46 ·25 cm, 5 lm column (Vydac). The conditions used
were: solvent A was water containing 0.1% trifluoroacetic
acid, solvent B was acetonitrile containing 0.1% trifluoro-
acetic acid and the gradient was 25–55% solvent B in 60 min.
Details regarding the PCI purification protocol have been
published elsewhere [20]. ProNtPCI mutant proteins used in
the in vivo refolding experiments were directly analyzed
by RP-HPLC on a Nova-Pak C8 3.9 ·150 mm column
(Waters), after sample acidification with trifluoroacetic acid
and filtration through 4 mm, 0.2 lm poly(vinylidene diflu-
oride) filters (National Scientific).
For protein production in E. coli BL21(DE3) cells, the
cultures were grown until they reached a value of 1
attenuation unit at 550nm, induced by addition of 0.2 m
M
isopropyl thio-b-
D
-galactoside, and cells were harvested by
centrifugation 2.5 h after induction. The cell pellet from a
1 L culture was resuspended in 50 mL 20 m
M
Tris/HCl,
0.5 m
M
EDTA (pH 8.5) and was maintained on ice for
10 min. The solution was sonicated for 10 min on ice at
50 Hz at half power, on a Labsonic-Braun sonicator and
centrifuged at 22 000 gfor 25 min. The pellet was resus-
pended in 50 mL 20 m
M
Tris/HCl, 0.5 m
M
EDTA and 2%
Triton X-100 (pH 8.5) and centrifuged at 22 000 gfor
25 min. The pellet was resuspended in 10 mL 6
M
guani-
dinium chloride and 30 m
M
dithiothreitol (pH 8.5). After
6 h the sample was centrifuged at 3000 gfor 10 min and
the supernatant was dialyzed against 0.1
M
Tris/HCl
(pH 8.5) for 12 h and then renaturation was performed
by dialysis in the presence of a redox system containing
4m
M
Cys and 2 m
M
Cys-Cys (cystine) at pH 8.5 for 48 h
at 4 C with a 3500-Da cut-off membrane (Spectrum
Medical Industries Inc). After dialysis, the sample was
centrifuged at 3000 gfor 10 min and the supernatant was
purified by RP-HPLC, on a Protein C4 0.46 ·25 cm,
5lm column (Vydac). The peptide corresponding to the
N-terminal pro-sequence (ProNt) was obtained by solid-
phase chemical synthesis. The released peptide was purified
by RP-HPLC on a Protein C4 1 ·25 cm, 5 lm column
(Vydac), in a linear gradient 20–27% in 7 min and 27–40%
solvent B in 26 min.
In vitro
folding experiments
One hundred micrograms of lyophilized aliquots of PCI,
185 lg lyophilized aliquots of ProPCI and 171 lg lyophi-
lized aliquots of ProNtPCI were used in each folding
experiment. The proteins were dissolved in 0.5 mL Tris/HCl
(0.5
M
, pH 8.5) containing 5
M
guanidinium chloride and
30m
M
dithiothreitol, to a final protein concentration of
46.5 l
M
. After 2 h at 25 C, the reduced and denatured
proteins were passed through a PD-10 (Pharmacia) column
equilibrated with 0.1
M
Tris/HCl buffer (pH 8.5). The
proteins were eluted in 1.2 mL and split in three parts which
were diluted to a final protein concentration of 14.5 l
M
,
with the 0.1
M
Tris/HCl buffer (pH 8.5), the same buffer
containing 1 m
M
Cys and the same buffer containing 4 m
M
Cys and 2 m
M
Cys-Cys, respectively. In the experiments
where the N-terminal pro-sequence was tested in trans,the
peptide was added to the denatured and reduced PCI in the
dilution buffer, to a final concentration of 14.5 l
M
.Samples
3642 S. Bronsoms et al. (Eur. J. Biochem. 270)FEBS 2003

of all reaction mixtures were collected in a time-course
manner for up to 24 h and trapped by mixing with an equal
volume of: (a) 1% trifluoroacetic acid in water (reversible
trapping) followed by analysis by RP-HPLC on a Protein
C4 0.46 ·25 cm, 5 lm column (Vydac). The gradient was
linear: 20–40% solvent B in 30 min for PCI, 25–35%
solvent B in 10 min and 35–45% solvent B in 40 min for
ProNtPCI and 20–30% solvent B in 5 min and 30–50%
solvent B in 30 min for ProPCI; (b) 0.1
M
iodoacetic acid
in Tris/HCl buffer (0.5
M
, pH 6.5) containing 40% (by
volume) of dimethylformamide (irreversible trapping) [25].
Carboxymethylation was allowed to proceed for 30 min at
25 C.
Inhibitory activity
The substrate used to perform the carboxypeptidase
activity was 0.2 m
M
furyl-acryloyl-
L
-phenylalanyl-
L
-phenyl-
alanine and the buffer was 50 m
M
Tris/HCl, 0.5
M
NaCl,
pH 7.5. To 985 lL of substrate, 5 lL of bovine
carboxypeptidase A (CPA) (Sigma) at 0.02 mgÆmL
)1
were added and the absorbance change at 330 nm was
measured during 2 min; then 10 lLofincreasing
concentrations of PCI or ProNtPCI were added and
the measures were continued for 2 min. The slope of the
first part of the assay corresponded to m
o
and the slope
of the second part to m
i
. The residual enzymatic activity
was calculated (m
o
to m
i
) and plotted as function of the
inhibitor concentration.
Mass spectrometry
Molecular mass was determined by MALDI-TOF mass
spectrometry on a Bruker–Biflex spectrometer. Ionization
wasaccomplishedwitha337-nmpulsednitrogenlaser
and spectra were acquired in the linear positive ion mode,
using a 19 kV acceleration voltage. Samples were pre-
pared mixing equal volumes of the protein solution and a
saturated solution of sinapinic acid, used as a matrix, in
aqueous 30% acetonitrile with 0.1% trifluoroacetic acid
(v/v).
Circular dichroism spectroscopy
CD spectra were collected on a Jasco-J715 spectropolari-
meter at 25 C, using a 2-mm path length cell, a band width
of 2 nm, a step size of 0.5 nm and an averaging time of 1 s.
Samples were analyzed in 0.1% trifluoroacetic acid (pH 2.0)
or 50 m
M
Na
2
HPO
4
(pH 7.0), at 100 lgÆmL
)1
final con-
centration.
Deuterium to proton (D/H) exchange
Fifteen micrograms of lyophilized samples of PCI or
ProNtPCI were resuspended in 5 lLofD
2
O and incubated
for 3 h at 50 C in order to exchange completely all labile
protons and afterwards were maintained for 30 min at
room temperature to refold properly. The native deuterated
proteins were diluted with four volumes of 15 m
M
glycine
pH 3.0 in H
2
O to start the hydrogen exchange and samples
were taken in a time-course manner and analyzed by
MALDI-TOF MS.
Exoproteolysis
Fifteen micrograms lyophilized aliquots of ProNtPCI were
dissolved in 10 lL10m
M
Tris/HCl buffer (pH 8.5) con-
taining 5 lg of leucine aminopeptidase (Sigma). Samples
were collected in a time-course manner, diluted with water
containing 0.1% trifluoroacetic acid (1 : 2) and the pro-
teolyzed products present in the mixture were identified
by MALDI-TOF mass spectrometry.
Nuclear magnetic resonance
NMR spectra were recorded on a Bruker AMX spectro-
meter operating at 500 MHz. Two milligrams of PCI, 2 mg
of the N-terminal pro-sequence peptide and 100 lgof
ProNtPCI were resuspended in 500 lLofNaH
2
PO
4
pH 4.00 containing 10% D
2
O and the spectra were
acquired at 35 C.
Results
Expression in
E. coli
In order to study experimentally whether PCI pro-regions
influence the folding of mature PCI, two precursor forms of
PCI were obtained by recombinant expression. The first
expression trials of ProNtPCI in E. coli MC1061 using a
pINIIIOmpA3-derived secretion vector led to a low yield of
purified protein due to the proteolysis of the pro-sequence
during the protein expression period. The ProNtPCI protein
wasdegradedtoPCIthataccumulatedintheculture
medium. The final yield of intact ProNtPCI was very low
(50 lgÆL
)1
) and it was used exclusively for the experiments
requiring small amounts of protein. In another expression
system ProNtPCI was cloned into pBAT4 vector with and
without the signal peptide OmpA, to produce the protein
either extracellularly or intracellularly in BL21 cells. The
extracellular expression of the molecule resembled that of
ProNtPCI in MC1061 cells. The intracellular expression led
to the formation of inclusion bodies, probably due to the
fact that PCI contains three disulfide bonds that can not be
efficiently formed inside the reductive environment of the
host cells, leading to the accumulation of protein aggregates.
After purification and disaggregation of the inclusion
bodies, the final yield of ProNtPCI produced with this
system was 3.4 mgÆL
)1
(see Experimental procedures). The
same expression protocol was used for the intracellular
expression of ProPCI, the other precursor form analyzed in
the refolding studies, which gave a final yield of 3.0 mgÆL
)1
of protein.
Refolding
in vitro
Mature PCI refolding undergoes a two-stage process: a first
stage of fast unspecific disulfide formation is followed by
a second stage (rate limiting step) of disulfide reshuffling
which leads to the native form [18]. Such behaviour was
investigated for the different recombinant molecular forms
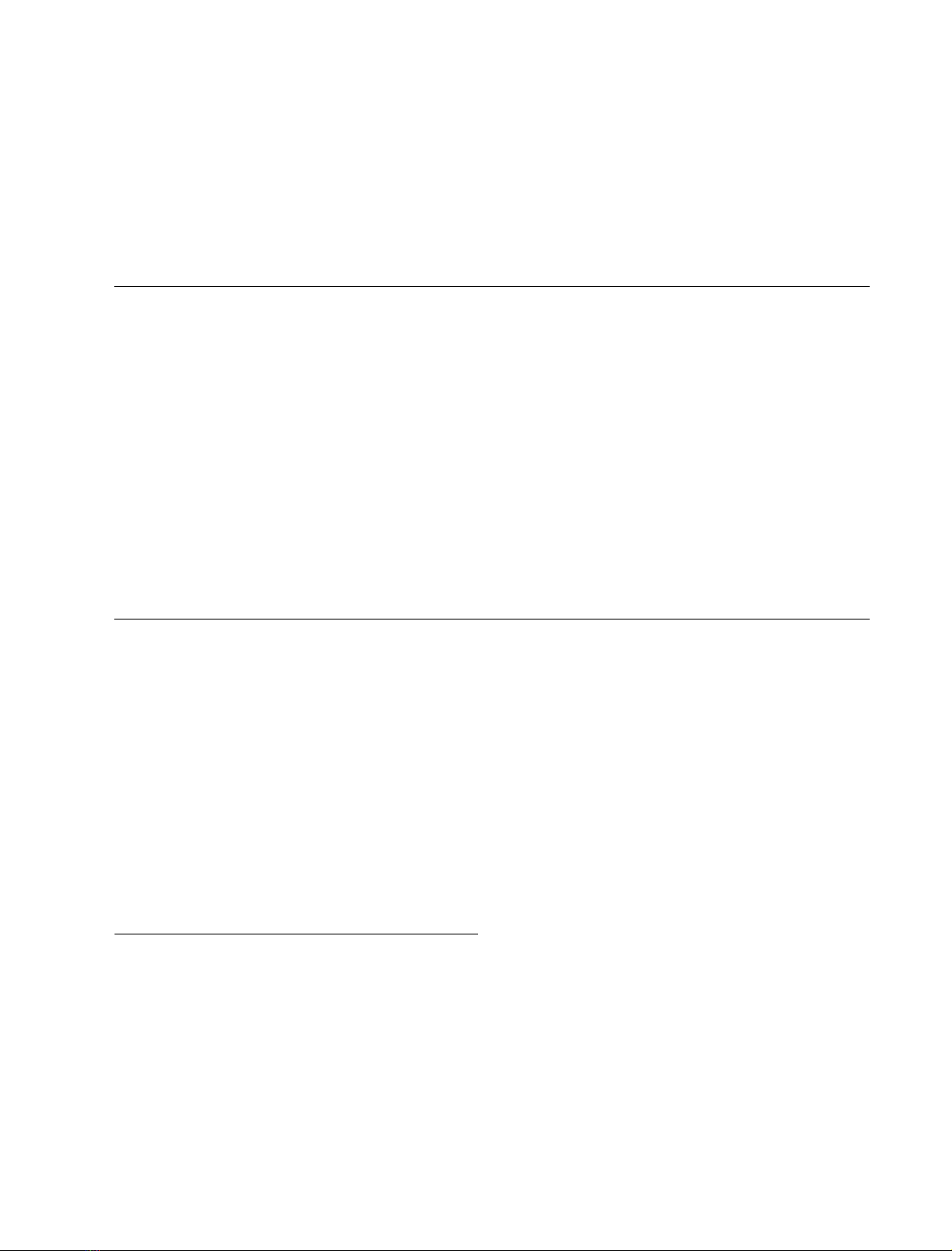
of this study. The yield of native-like forms achieved after 7 h
of refolding in absence of an external thiol was similar in all
the molecules tested (<5%) (Fig. 1A, left). The RP-HPLC
chromatogram profiles from the 7 h refolding mixture of
FEBS 2003 Folding of PCI precursor form (Eur. J. Biochem. 270) 3643
PCIandPCIplustheProNtintrans were indistinguishable.
Thus, we can assume that the molar ratio among scrambled
and native species is not affected by the addition of the ProNt
segment in trans. The ratio between the native form and the
ensemble of scrambled species remains constant for all tested
forms. Therefore, in the absence of redox agents, neither the
N-terminal nor the C-terminal PCI pro-sequences have an
effect on the final yield of native PCI, indicating that the
overall folding process is similar among all the molecules
tested under these conditions.
It is worth mentioning that the folding of PCI is
accelerated by the presence of external thiols in the folding
mixture [18]. While the addition of Cys accelerates the
second stage of disulfide reshuffling of scrambled forms to
native PCI, the addition of Cys-Cys enhances the first
stage of disulfide formation, which leads to the formation
of scrambled species. We analyzed the refolding process of
all the above mentioned recombinant forms in the
presence of 4 m
M
Cys and 2 m
M
Cys-Cys. The RP-HPLC
profiles show that the folding kinetics and the fold-
ing efficiency are higher under such conditions. The
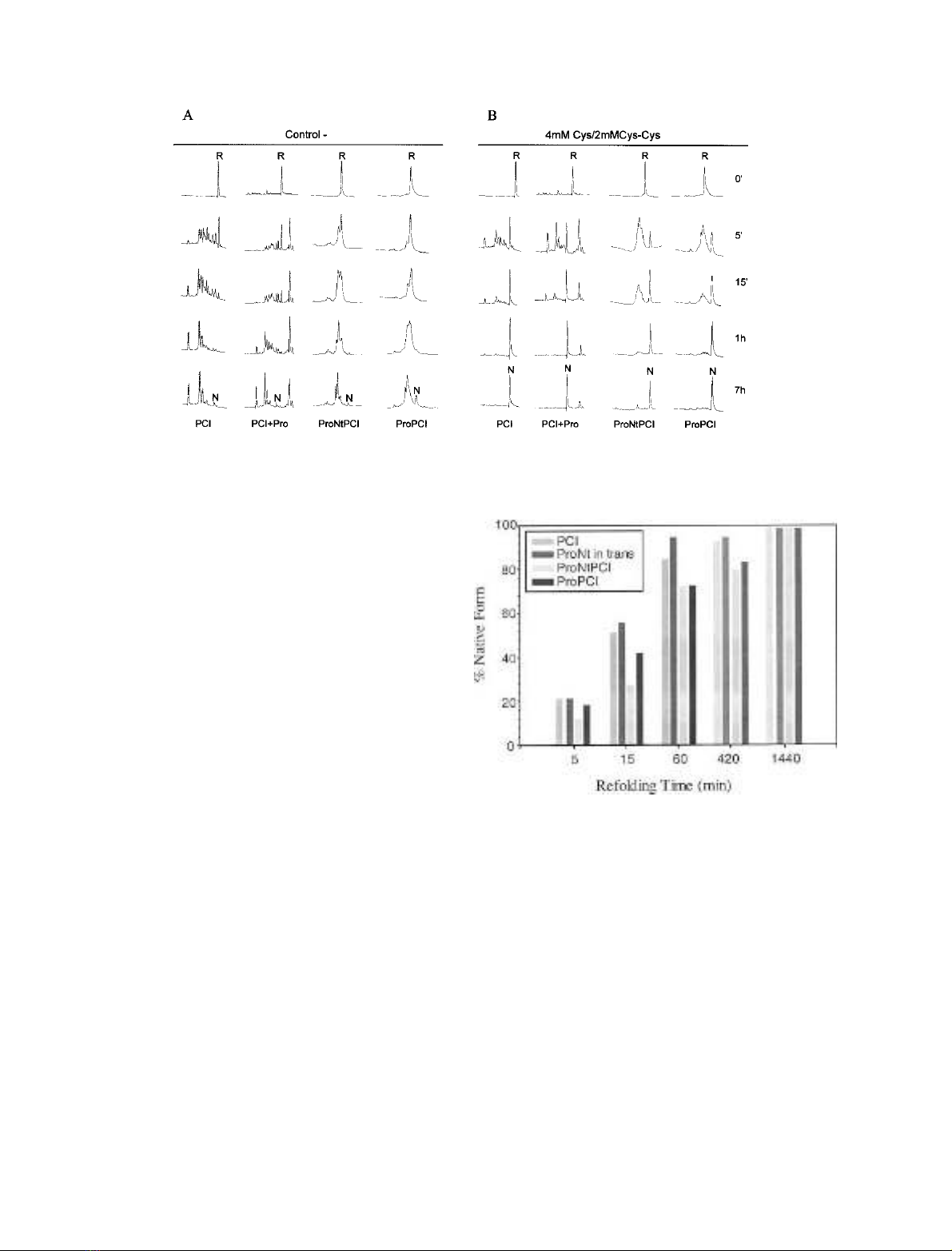
yield of native species achieved is superior to 70% after
1 h of refolding (Fig. 1B). The folding kinetics and
efficiency of PCI and PCI plus ProNt peptide in trans
are similar but, surprisingly, for ProNtPCI and ProPCI
the folding kinetics are a little slower (Figs 1B and 2).
Nevertheless, they can be considered to have a similar
folding efficiency, as the final yields of native form at 24 h
of refolding are nearly identical (Fig. 2). The flow of
intermediate species containing one, two and three
disulfides was followed by MALDI-TOF mass spectro-
metry analyzing the iodoacetate-trapped folding inter-
mediates in the four sets of tested molecules. The flow of
refolding intermediates is characterized by a progression
from the reduced state through the more thermodynam-
ically stable 1-, 2- and 3-disulfide species. The rate of
disulfide formation was similar in all the molecules tested
under the same conditions.
Influence of pro-sequences
in vivo
It has been reported that some mutations of mature PCI at
the C-tail give rise to low expression yields or low folding
efficiencies compared to wild-type mature PCI: D3, Y37G
and G35P/P36G PCI [22]. To determine whether the
N-terminal pro-region might improve their in vivo expres-
sion or folding in E. coli, the expression of each PCI mutant
protein and wild-type PCI was analyzed in parallel to the
corresponding ProNtPCI mutant protein and wild-type
ProNtPCI. Twenty-four hours after induction of protein
expression the supernatant was collected, analyzed by
RP-HPLC (Fig. 3) and the species were identified by
Fig. 1. In vitro folding studies of PCI, PCI plus the ProNt in trans, ProNtPCI, and ProPCI in the presence of selected redox agents. Reduced and
denatured proteins were refolded in the absence (no added thiols) (A), or presence (4 m
M
Cys/2 m
M
Cys-Cys) (B) of external thiols. Folding
intermediates were acid-trapped and analyzed by RP-HPLC. Elution positions of native (N) and reduced (R) forms are indicated.
Fig. 2. Refolding efficiencies of PCI, PCI plus the ProNt peptide in
trans,ProNtPCIandProPCI.Reduced and denatured proteins were
refolded in the presence of 4 m
M
Cys/2 m
M
Cys-Cys and acid-trapped
folding intermediates were analyzed by RP-HPLC. The yield of native
form was calculated in each time point from the peak areas in the
corresponding RP-HPLC chromatograms.
3644 S. Bronsoms et al. (Eur. J. Biochem. 270)FEBS 2003

MALDI-TOF mass spectrometry. As previously mentioned,
theN-terminalpro-regionisdegradedintheE. coli
extracellular media when secreted therefore the protein
species found in the culture media were the mature PCI
forms without the pro-region.
The amount of each expressed protein was calculated by
comparison of the corresponding RP-HPLC peak areas
(data not shown). The final yield of each native-like
ProNtPCI mutant, the ratio between the native form and
the ensemble of scrambled species, and the ratio among the
scrambled species present in the cell culture supernatants
were compared with those of PCI mutants to evaluate any
influence of the pro-region. Under the conditions of the
experiment, the values obtained were similar for mature PCI
and the ProNtPCI mutant proteins, indicating that the pro-
region of PCI affects neither the expression levels nor the
folding efficiencies in vivo in E. coli.
Inhibitory activity
To test whether ProNtPCI displays the same biological
activity as mature PCI, inhibition studies of carboxypep-
tidase A1 (CPA1) enzyme by ProNtPCI and PCI were
performed. Both proteins show very similar affinities for
CPA1 using the substrate furyl-acryloyl-
L
-phenylalanyl-
L
-phenylalanine and they have the same IC
50
value (100 n
M
).
According to these results, the mature PCI region within
ProNtPCI should keep the same disulfide pairing and a
similar three-dimensional structure as in isolated mature
PCI, at least in the region which docks with the enzyme.
Structural analyses
Despite the small amounts of regular secondary structures
of wild-type PCI native form [26], far-UV CD spectroscopy
may be helpful to indicate its folding state, as it shows a
characteristic positive ellipticity band at 228 nm when it is
properly folded and possesses the wild-type Y37 residue
[22]. Thus, this maximum band at 228 nm would also be
expected for ProNtPCI. However, when the CD spectrum
of ProNtPCI was recorded, such a spectral band was not
observed at pHs of either 2.0 or 7.0 (Fig. 4). So, apparently,
the environment of Y37 is affected in the pro form.
Time-course deuteron to proton exchange monitored
by MALDI-TOF MS [27] was also performed for both
proteins. PCI contains 65 labile hydrogens and NMR has
demonstrated that five of them form the slow exchange core
[26]. The results indicate that the hydrogen exchange
kinetics followed by both proteins are similar (Fig. 5). For
each protein a major subpopulation of protons exchange
rapidly (within 2 h) and the equilibrium is reached after
24 h. However, the number of slow exchanging deuterons is
significantly different. While PCI retains five deuterons
protected from exchange when equilibrium is reached,
ProNtPCI retains nine under the same conditions. In
addition, the number of deuterons retained before achieving
Fig. 3. In vivo expression of recombinant forms of wild-type PCI, wild-type ProNtPCI and variants of them with mutations at the C-tail. (A) Schematic
representation of the recombinant proteins produced for this study. Amino acids are in one-letter code. The N-terminal pro-sequence is indicated
with a white box, the mature protein with a light shaded box and the mutated amino acids with a white box. (B) Recombinant proteins were
produced in E. coli MC1061 cells in the expression vector pINIII-OmpA3. The supernatants, after 24 h induction, were analyzed by RP-HPLC.
The quantity of each native and scrambled form was calculated from the peak area of its corresponding RP-HPLC chromatogram. The elution
position of each native or native-like form is indicated (N). In case of G35P/P36G mutants the disulfide pairing is not the same as wild-type PCI [22];
in these cases, S stands for the most stable form.
FEBS 2003 Folding of PCI precursor form (Eur. J. Biochem. 270) 3645


























