
The thermodynamic analysis of protein stabilization by sucrose
and glycerol against pressure-induced unfolding
The typical example of the 33-kDa protein from spinach photosystem II
Kangcheng Ruan
1
, Chunhe Xu
2
, Tingting Li
1
, Jiong Li
1
, Reinhard Lange
3
and Claude Balny
3
1
Laboratory of Proteomics, Institute of Biochemistry and Cell Biology, Shanghai Institute for Biological Science,
Chinese Academy of Sciences, Shanghai, China;
2
Institute of Plant Physiology, Shanghai Institute for Biological Science,
Chinese Academy of Sciences, Shanghai, China;
3
Institut National de la Sante
´et de la Recherche Me
´dicale,
INSERM U 128, IFR 24, CNRS, Montpellier, France
We have studied the reaction native «denatured for the
33-kDa protein isolated from photosystem II. Sucrose
and glycerol have profound effects on pressure-induced
unfolding. The additives shift the equilibrium to the left;
they also cause a significant decrease in the standard
volume change (DV). The change in DVwas related to
the sucrose and glycerol concentrations. The decrease in
DVvaried with the additive: sucrose caused the largest
effect, glycerol the smallest. The theoretical shift of the
half-unfolding pressure (P
1/2
)calculatedfromthenet
increase in free energy by addition of sucrose and glycerol
was lower than that obtained from experimental mea-
surements. This indicates that the free energy change
caused by preferential hydration of the protein is not the
unique factor involved in the protein stabilization.
The reduction in DVshowed a large contribution to the
theoretical P
1/2
shift, suggesting that the DVchange,
caused by the sucrose or glycerol was associated with the
protein stabilization. The origin of the DVchange is
discussed. The rate of pressure-induced unfolding in the
presence of sucrose or glycerol was slower than the
refolding rate although both were significantly slower
than that observed without any stabilizers.
Keywords: conformational changes; hydrostatic pressure;
spinach particle; protein denaturation.
Understanding protein folding mechanisms is one of the big
challenges in protein science. For example, an unusual
property of prion protein unfolding in neutral salt solution
has recently been shown [1]. However, the prion protein is
not easy to work with and to go further, convenient models
must be used. The 33-kDa protein from spinach photosys-
tem II is a good system with which to explore the role of
additives in protein folding and unfolding; their effects on
the chemical denaturation of this protein have been
described previously. This protein has a very low free
energy of unfolding and it is easy to modulate its unfolding
transition [2].
Most protein denaturation studies use chemicals (such as
urea or guanidine hydrochloride) or thermal perturbation to
influence the folding pattern. Reversibility is frequently a
problem. For many years, various chemicals like neutral
salts, glycerol, sucrose have been known as protein stabi-
lizers. Initially it was thought that these molecules could
form coating shells around the proteins. Subsequently, other
studies on sucrose and glycerol indicated that these
substances do not usually bind to protein; their presence
changes the water surface tension around protein. They are
preferentially depleted from the protein surface layer [3–5].
In other words, the proteins are preferentially hydrated
around the surface in the presence of these stabilizers. This
leads to an increase in free energy and consequently
protection against denaturation [5].
An increasing number of researchers are using high-
pressure as a denaturing agent. Compared to other meth-
ods, pressure denaturation is often rapidly reversible [6].
High hydrostatic pressure has been used extensively to
denature single chain proteins and oligomeric proteins
[6–17]. Generally, single chain proteins such as trypsin,
chymotrypsinogen, phospholipase, etc. can be unfolded in
the pressure range 300–600 MPa; the 33-kDa protein and
staphylococcal nuclease unfold at lower pressures [10,11].
High pressure induces a system volume decrease which
governs the protein unfolding equilibrium; it has been
shown that this volume change can be modulated by various
factors. Different workers have studied this phenomenon
[10,12,18]
1. For example, Royer and coworkers found that
Correspondence to K. Ruan, Shanghai Institute of Biochemistry and
Cell Biology, Chinese Academy of Sciences, 320 Yue-Yang Road,
Shanghai 200031, China. Fax: + 86 21 64338357,
Tel.: + 86 21 64740532, E-mail: kcruan@sunm.shcnc.ac.cn or
C. Balny, INSERM U128, 1919 route de Mende, 34293 Montpellier
Cedex 5, France. Fax: +33 47523681, Tel.: +33 467613360,
E-mail: balny@montp.inserm.fr
Abbreviations: GdmCl, guanidinium chloride; 4thD, fourth derivative
absorbance spectra; CSM, centre of spectral mass; P
1/2
, experimental
half pressure of denaturation; P
1/2
*, value of half pressure
denaturation obtained from calculation for the net increase in free
energy; P
1/2
**, value of half pressure denaturation obtained from
calculation for the net reduction of the standard volume change.
(Received 7 October 2002, revised 9 December 2002,
accepted 27 January 2003)
Eur. J. Biochem. 270, 1654–1661 (2003) FEBS 2003 doi:10.1046/j.1432-1033.2003.03485.x

xylose stabilizes staphylococcal nuclease mainly by increas-
ing the protein free energy (DG) of denaturation, while the
standard volume change (DV) seems to be independent of
the xylose concentration [10,18].
In earlier work we explored the pressure-induced dena-
turation of the 33-kDa protein isolated from spinach
photosystem II; the equilibrium is a two-state reversible
transition which is influenced by NaCl [11]. P
1/2
(the half-
pressure of denaturation) was shifted from 118 MPa to
127 MPa and 195 MPa in the absence and in the presence of
0.5
M
and 1.0
M
NaCl, respectively. It was also observed that
the volume change, DV, decreases from )120.0 (without salt)
to )108.1 (0.5
M
NaCl) and to )80.0 mLmol
)1
(1.0
M
NaCl). DVand DG both contribute to the folding–unfolding
shift. Some questions are still without clear answers: (a) is the
reduction in DValso found with stabilizing agents such as
glycerol and sucrose? (b) with these agents, is the same
stabilization mechanism involved when the denaturation is
induced either by chemical denaturants or by hydrostatic
pressure?
To answer these questions the effect of sucrose and
glycerol on the pressure-induced unfolding of the 33-kDa
protein has been studied in the present work.
We chose the 33-kDa protein as a model because of its
very low DG of unfolding at pH 6.0 and 20 C()3.5
kcalÆmol
)1
) and because of its large standard DV
()120 mLÆmol
)1
). The response of the 33-kDa protein
to pressure is completely reversible [11]. Moreover, the
protein molecule contains only one tryptophan residue
(Trp241) buried in a very strong hydrophobic region.
This allows for easy fluorescence detection when this
residue is exposed to the solvent. In the native form, the
fluorescence emission, k
max
, is at 317 nm, shifting to
352 nm when unfolded.
In this report we show that sucrose, glycerol and NaCl
protect the 33 kDa protein against denaturation by either
hydrostatic pressure or guanidine hydrochloride.
Materials and methods
Purification of the 33-kDa protein
The 33-kDa protein was isolated and purified from
spinach chloroplast photosystem II as described in our
previous report [11]. The purified protein was dialysed
against 10 m
M
NH
4
HCO
3
and then lyophilized. The
protein concentrations were determined as described by
Xu and Briker [19]. In most experiments, the protein was
dissolvedin0.05
M
pH 6.0 Mes buffer. All other reagents
were of A. R. grade. Distilled water was further purified
by a Millipore system to a resistance of 18 MW.
Fluorescence measurements
The fluorescence measurements were carried out either on
an Aminco Bowman Series 2 (AB2) fluorospectrophoto-
meter (SLM Co.) or on a SLM 48000 fluorospectrophoto-
meter (SLM Co.). These have been modified thereby
allowing us to measure fluorescence in a pressure range
from 0.1 MPa to 600 MPa at temperatures between )20 C
and 100 C. The fluorescence spectra were quantified by
specifying the centre of spectral mass (CSM) <m>as
introduced in our previous and related papers [20,21]. The
excitation wavelength for the intrinsic fluorescence was
295 nm, which excited only the tryptophan residue.
To measure the unfolding–refolding kinetics of the
protein, the fluorescence spectrophotometer was further
modified to adapt a pressure jump device designed in the
INSERM laboratory [22]. Positive or negative pressure-
jumps up to 150 MPa were possible in a pressure range
from 0.1 to 600 MPa, with a dead time of 5 ms.
Fourth derivative UV absorbance spectra
Absorption spectra of the protein between 260 and 300 nm
were recorded at 20 C using a modified Cary3 (Varian)
absorption spectrophotometer as described elsewhere; this
instrument allows experiments in a pressure range from
atmospheric pressure to 500 MPa at temperatures between
)20 Cand100C [23]. The 4th derivative (4
th
D) absorb-
ance spectra were calculated from the corresponding
absorption spectra as described previously [23,24].
Unfolding degree calculations
The basic scheme for a denaturation reaction is N «D
where N and D are the native and the denatured forms,
respectively. The method for determining the degree of
unfolding of the protein (a) was the same as reported
previously and was calculated either from the centre of
spectral mass (CSM) <m> for fluorescence measurement
or from the amplitude of the change at 293 nm in the 4thD
spectra [11]. The degree of unfolding (a) was plotted against
pressure to draw the unfolding curve and to determine the
half-denaturation pressure, P
1/2
. The free energy and
standard volume change were calculated from the unfolding
curve according to the method of Li et al. [13]. The values of
DG were also estimated from half-denaturation pressure,
P
1/2
, according to:
DG¼0:234 DVP1=2
where P
1/2
is in MPa, DVin mLÆmol
)1
and DGincalÆmol
)1
,
respectively [11].
The free energy of unfolding due to guanidine hydro-
chloride was calculated according to the Tanford method
[2,25].
Results
Sucrose stabilization of the 33-kDa protein
pressure-induced unfolding
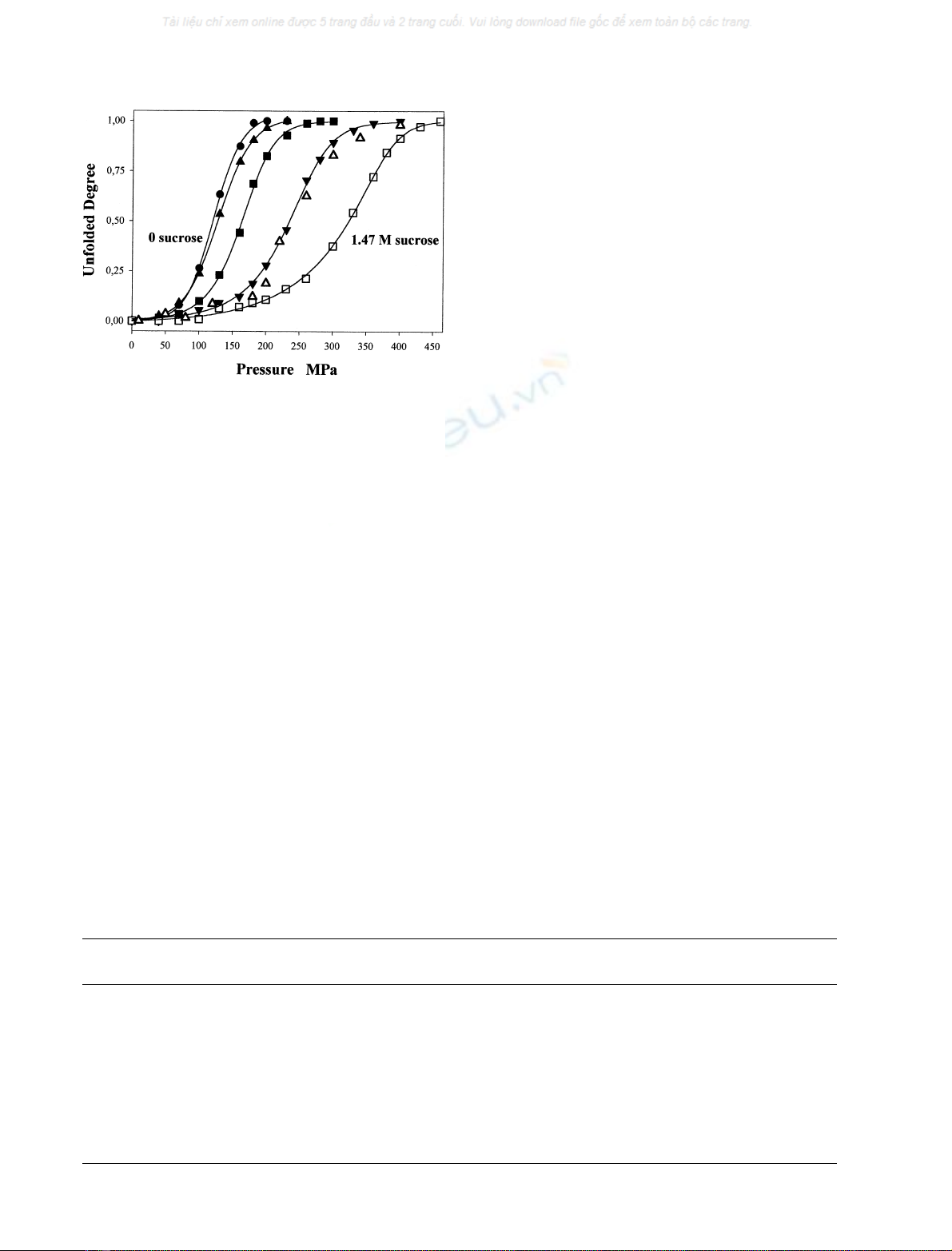
Fig. 1 shows the degree of unfolding, a,ofthe33-kDa
protein plotted against pressure. The curves are shifted to
the higher pressures as the sucrose concentration is
increased. Consequently, P
1/2
is shifted from a minimum
of 118 MPa to 320 MPa at 1.47
M
sucrose. This indicates
that in the presence of sucrose, the 33-kDa protein is more
stable and is protected from pressure-induced denaturation.
DG of unfolding is listed in Table 1. It increases as the
sucrose concentration increases. This observation is in good
agreement with the Timasheff model and with the results
reported by Frye and Royer for the xylose study on
FEBS 2003 Stabilization of 33-kDa protein of spinach PS II against pressure unfolding (Eur. J. Biochem. 270) 1655
staphylococcal nuclease [18]. In contrast to the free energy,
DVis found to decrease with the sucrose addition. In the
absence of sucrose, DVis )120 mLÆmol
)1
; it decreases to
53.7 mLÆmol
)1
at 1.47
M
sucrose. The decrease in DVis
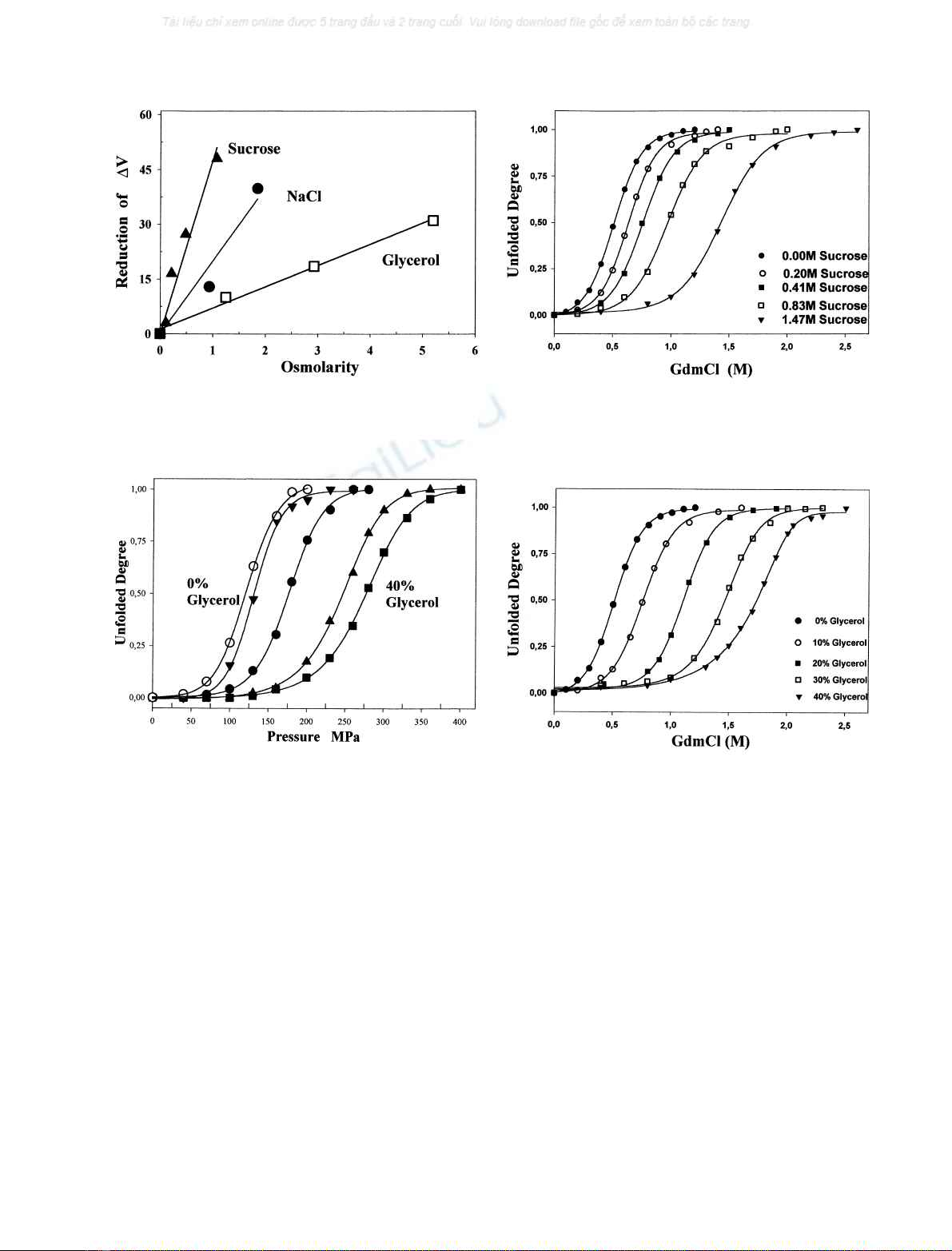
obviously dependent on the sucrose concentration. Fig. 2
shows that the change in DVreduction is a linear function of
the sucrose concentration (in osmolarity). However, the
linearity is not followed when the concentration is rather
high (1.47
M
).
DGandDVof unfolding in the presence of sucrose have
been also determined using 4thD spectra. The unfolding
curve of the protein in the presence of 0.83
M
sucrose
obtained from the 4thD spectra (n)asshowninFig.1is
very close to that obtained from the tryptophan fluorescence
measurement (m). The free energy and the standard volume
change are 3.94 kcalÆmol
)1
and )66.1 mLÆmol
)1
, in good
agreement with that obtained from fluorescence experi-
ments (3.9 kcalÆmol
)1
and )72 mLÆmol
)1
).
Stabilization effect of glycerol
Fig. 3 shows the effect of glycerol on pressure-induced
unfolding of the 33-kDa protein. When the glycerol
concentration is increased from 0 to 40%, the unfolding
curves shift to the higher pressures. P
1/2
increases from
118 MPa to 280 MPa (see Table 1). In 40% glycerol the
33-kDa protein is totally unfolded at about 400 MPa, a
value much higher than that observed in the absence of
glycerol (180 MPa). These results indicate that the glycerol,
like sucrose, stabilizes the protein against pressure denatur-
ation. DG of unfolding in the presence of glycerol increases
from 3.5 to 5.3 kcalÆmol
)1
(see Table 1); DG is dependent
on glycerol concentration. All of these results also provide
evidence supporting the Timasheff model [5]. DVof
unfolding decreases with increasing glycerol concentration
(see Table 1). It goes from )120 mLÆmol
)1
(without
glycerol) to )80.4 mLÆmol
)1
(in 40%), giving results similar
to those obtained from experiments using sucrose. Import-
antly the decrease in DVseems to be associated with the
stabilization effect. It should be noticed that the 10%
glycerol concentration is an exception. Under this condition
the DVhas a small increase (of 8mLÆmol
)1
), which is
similar to that observed in the staphylococcal nuclease study
[18]. They found a small increase in DVwhen xylose was
added. However, they found that the increase in DVis
independent of the xylose concentration. The linearity
between the reduction in DVand the glycerol concentration
isshowninFig.2(d).
Denaturation of the 33-kDa protein by guanidine
hydrochloride
To understand the sucrose and glycerol effects on the
stabilization of this protein against pressure-induced
denaturation, guanidinium chloride (GdmCl)-induced
protein denaturation has been studied. Tryptophan fluor-
escence was used as a probe. The unfolding curves are
plottedinFigs4and5forsucroseandglyceroleffects,
respectively. The unfolding curves are obviously shifted to
higher GdmCl concentrations when sucrose or glycerol
concentrations are increased. The free energy values are
listed in Table 1 and show a significant increase with the
sucrose or glycerol concentrations. This indicates that
Fig. 1. The unfolding of the 33-kDa protein induced by hydrostatic
pressure in the presence of different sucrose concentrations. Curves from
left to right: 0.0, 0.1, 0.2, 0.41, 0.83 and 1.47
M
sucrose, respectively.
The unfolding degrees (a) were calculated from the fluorescence
spectra of the protein excited at 295 nm or from the 4
th
Dspectrum.
Protein concentration for fluorescence and 4
th
D measurements:
0.1 mgÆmL
)1
and 0.7 mgÆmL
)1
, respectively, in 0.05
M
Mes buffer,
pH 6.0, 20 C.
Table 1. Thermodynamic parameters for the 33-kDa protein unfolding. DG, obtained from pressure-induced unfolding; DG*, obtained from GdmCl-
induced unfolding; TP
1/2
*, obtained from calculation for the net increase in free energy; TP
1/2
** obtained from calculation for the net reduction of
the standard volume change. Reactions were performed at pH 6.0 and 20 C.
DG
(kcalÆmol
)1
)
DV
(mLÆmol
)1
)
P
1/2
(MPa)
DG*
(kcalÆmol
)1
)
TP
1/2
*
(MPa)
TP
1/2
**
(MPa)
No sucrose 3.5 )120.0 118 )2.6 118 118
0.10
M
sucrose 3.4 )118.9 118 – 115 119
0.20
M
sucrose 3.2 )103.5 130 )3.1 109 136
0.41
M
sucrose 3.6 )92.7 163 )3.3 121 152
0.83
M
sucrose 3.9 )72.0 234 )3.8 132 197
1.47
M
sucrose 4.0 )53.7 320 )4.6 135 264
10% glycerol 4.0 )128.3 132 )3.3 135 110
20% glycerol 4.3 )101.5 177 )4.7 144 140
30% glycerol 5.2 )89.0 248 )5.2 175 159
40% glycerol 5.3 )80.4 280 )5.7 178 176
1656 K. Ruan et al.(Eur. J. Biochem. 270)FEBS 2003
both sucrose or glycerol can inhibit the chemical dena-
turation of the 33-kDa protein by the GdmCl, according
to the preferential hydration around protein surface in
the presence of the stabilizers. The values of DG
obtained from either pressure- or GdmCl-induced unfold-
ing are very similar. Some differences in quantitative
values are observed (but they remain within a reasonable
range for the results collected from various experimental
methods). DGforthenative«denatured transition in
the absence of protectants is 2.6 kcalÆmol
)1
, a value in
good agreement with those reported by Tanaka et al.
(2.8 kcalÆmol
)1
)[2].
The change in
P
1/2
is caused by effects on DG and D
V
P
1/2
, the pressure at which 50% of the protein is unfolded, is
a parameter often used to evaluate protein stability. The
higher is P
1/2
, the more stable is the protein to pressure-
induced denaturation. P
1/2
is related both to DGandDV
according to:
P1=2¼DG=DVor ln Kp ¼DGþPDV=RT
From the above formulae, the change in P
1/2
caused by the
net variation in DG or by the net standard change alone
(termed theoretical half unfolding pressure, TP
1/2
)canbe
obtained. The TP
1/2
caused both by the net increase in DG
and the net reduction of DVupon either sucrose or glycerol
addition were calculated and listed in Table 1. It was found
that the TP
1/2
* caused by the net increases in DG were lower
than the experimental P
1/2
. Typically, the difference between
P
1/2
and TP
1/2
*isaslargeas185 MPa in the presence of
1.47
M
sucrose and 102 MPa in 40% glycerol (see Table 1).
Even when the rather large values of DG obtained in
GdmCl denaturation experiments were used for calculation,
TP
1/2
*(153MPafor1.47
M
sucrose and 192 MPa for 40%
glycerol) were still much lower than those obtained from
Fig. 2. The effect of sucrose (m), glycerol (j)andsalt(d)onthe
standard volume change of the protein. The concentrations of the
additives are expressed in osmolarity.
Fig. 3. The unfolding of the 33-kDa protein induced by hydrostatic
pressure in the presence of different glycerol concentrations (in volume).
Curves from left to right: 0%, 10%, 20%, 30% and 40%, respectively.
Other conditions as in Fig. 1.
Fig. 4. The unfolding of the 33-kDa protein induced by GdmCl in the
presence of different sucrose concentrations. Other conditions as in
Fig. 1.
Fig. 5. The unfolding of the 33-kDa protein induced by GdmCl in the
presence of different glycerol concentrations (in volume). Other condi-
tionsasinFig.1.
FEBS 2003 Stabilization of 33-kDa protein of spinach PS II against pressure unfolding (Eur. J. Biochem. 270) 1657
measurements (320 MPa and 280 MPa, respectively). This
indicates that the net increase in free energy upon sucrose or
glycerol addition cannot explain totally the protection
effect—other factors are certainly involved. Meanwhile
it was found that the theoretical changes in TP
1/2
**
calculated from the net reduction of DVwere larger. In
thecaseofsucrose,theTP
1/2
** values calculated from the
DVmeasurements were larger than the values calculated
from the net increase in DG. Meanwhile, in the case of
glycerol the TP
1/2
** values calculated from both DVand
DG were close to each other. These observations indicate
that the reduction in DVprobably plays an important role in
the sucrose or glycerol protein protection against pressure
denaturation.
Effects of sucrose and glycerol on the kinetics of the
pressure induced protein unfolding and refolding
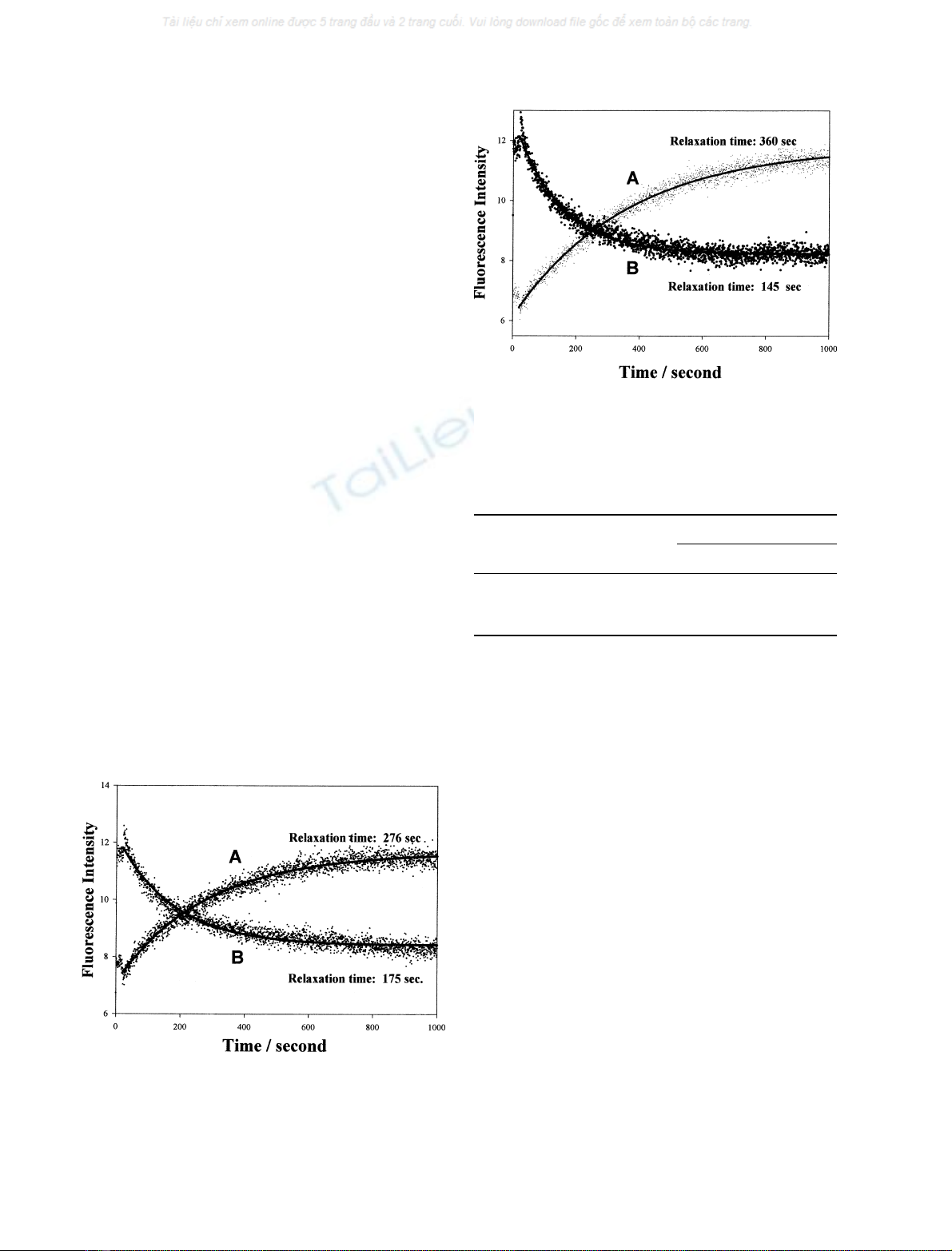
The unfolding and refolding kinetics in the presence of
either 0.83
M
sucrose or 30% glycerol have been investi-
gated using positive and negative pressure jumps (Figs 6
and 7). Fluorescence intensity was monitored at 350 nm
[11]. A single exponential was fit to the data (solid lines); the
curves showed a rather slow, two-state transition processes.
The corresponding relaxation times for the protein unfolding
and refolding in the presence of both 0.83
M
sucrose and
30% glycerol are listed in Table 2. Compared with the
kinetics of the protein unfolding–refolding measured with-
out additive where both are about 100 s [11], sucrose and
glycerol slow both the folding and unfolding rates. It was
also found that the presence of sucrose or glycerol induced
an unfolding relaxation time significantly longer than that
for the refolding reaction. For sucrose, the unfolding
relaxation time is 2.5 times longer than refolding, and, for
glycerol it is 1.5 times longer. The actual data suggest that
the rather slow unfolding rate could be associated with the
sucrose and glycerol stabilization effect.
Discussion
Our goal is to understand how high hydrostatic pressure
induces the refolding of proteins and how sugars and salts
influence this refolding. To this end, we used the effects of
both hydrostatic pressure and osmotic pressure as probes
[26]. As mentioned in the introduction, the stabilization
mechanism of these agents has been attributed to a protein
preferential hydration mechanism as proposed by Timasheff
[5] or by an osmotic stress [27] where, mathematically, the
two mechanisms cannot be distinguished [28].
In a very well documented paper, Parsegian et al.[28]
indicated that there has been much confusion about the
relative merits of different approaches, osmotic stress,
preferential interaction (i.e. preferential hydration), and
crowding, to describe the indirect effect of solutes on
macromolecular conformations and reactions. The two first
mechanisms (and crowding) cannot be distinguished as they
are derived from the same solution theory. In the prefer-
ential hydration model proposed by Timasheff, both the
chemical nature and the size of the solute determine water
exclusion from the protein surfaces [5]. Osmotic stress
emphasizes the role of the water that is necessarily included
if solutes are excluded [28], dealing also with the movement
of water molecules [27].
Upon addition of solutes (in the present case, stabilizers),
surface tension around the protein changes because of water
exclusion. Consequently, the protein free energy increases,
resulting in protein stabilization. However, the question is
Table 2. Relaxation time of unfolding and refolding of the protein
induced by pressure (times obtained from the data in Figs 6 and 7).
Sample condition
Relaxation time (s)
Unfolding Folding
0% Glycerol, 0
M
sucrose 125 100
30% Glycerol 360 145
0.8
M
Sucrose 276 175
Fig. 6. Kinetics of the pressure-induced unfolding and refolding of the
33-kDa protein in the presence of 0.83
M
sucrose. Curve A for a pressure
jump from 100 MPa to 180 MPa. Curve B for a pressure jump from
180 MPa to 100 MPa. Solid lines are the fitted curves. Protein con-
centration: 0.1 mgÆmL
)1
in 0.05
M
Mesbuffer,pH6.0,20C. Exci-
tation wavelength, 295 nm; emission wavelength, 350 nm.
Fig. 7. Kinetics of the pressure-induced unfolding and refolding of the
33-kDa protein in the presence of 30% glycerol. Otherconditionsasin
Fig. 6.
1658 K. Ruan et al.(Eur. J. Biochem. 270)FEBS 2003


























