
Cloning of a rat gene encoding the histo-blood group A enzyme
Tissue expression of the gene and of the A and B antigens
Anne Cailleau-Thomas
1
,Be
´atrice Le Moullac-Vaidye
1
,Je
´zabel Rocher,
1
Danie
`le Bouhours
2
,
Claude Szpirer
3
and Jacques Le Pendu
1
1
INSERM U419, Institut de Biologie, Nantes, France;
2
INSERM U539, Faculte
´de Me
´decine, Nantes Cedex, France;
3
IBMM, Universite
´Libre de Bruxelles, Gosselies, Belgium
The complete coding sequence of a BDIX rat gene homo-
logous to the human ABO gene was determined. Identifi-
cation of the exon–intron boundaries, obtained by
comparison of the coding sequence with rat genomic
sequences from data banks, revealed that the rat gene
structure is identical to that of the human ABO gene. It
localizes to rat chromosome 3 (q11-q12), a region homolo-
gous to human 9q34. Phylogenetic analysis of a set of
sequences available for the various members of the same
gene family confirmed that the rat sequence belongs to the
ABO gene cluster. The cDNA was transfected in CHO cells
alreadystablytransfectedwithana1,2fucosyltransferase in
order to express H oligosaccharide acceptors. Analysis of the
transfectants by flow cytometry indicated that A but not B
epitopes were synthesized. Direct assay of the enzyme
activity using 2¢fucosyllactose as acceptor confirmed the
strong UDP-GalNAc:Fuca1,2GalaGalNAc transferase
(A transferase) activity of the enzyme product and allowed
detection of a small UDP-Gal:Fuca1,2GalaGal transferase
(B transferase) activity. The presence of the mRNA and of
the A and B antigens was searched in various BDIX rat
tissues. There was a general good concordance between the
presence of the mRNA and that of the A antigen. Tissue
distributions of the A and B antigens in the homozygous
BDIX rat strain were largely different, indicating that these
antigens cannot be synthesized by alleles of the same gene in
this rat inbred strain.
Keywords:ABO;N-acetylgalactosaminyltransferase; histo-
blood group; antigen; rat.
Histo-blood group antigens A and B are oligosaccharides
carried by glycolipids and glycoproteins or present as free
oligosaccharides in some biological fluids such as milk or
urine. The immunodominant A and B epitopes correspond
to the trisaccharides GalNAca1,3(Fuca1,2)Galb-and
Gala1,3(Fuca1,2)Galb-, respectively. In humans, the ABO
gene is polymorphic with A alleles encoding A transferases,
B alleles encoding B transferases and O alleles encoding
inactive products. The A transferases catalyse the transfer of
an N-acetylgalactosamine to acceptor H substrates
(Fuca1,2Galb-) whereas the B transferases catalyse the
transfer of a galactose to the same substrates [1]. ABH
antigens are found in many species and have a wide tissue
distribution. Their main sites of expression appear to be
epithelia in contact with the external environment such as
the gut, the higher respiratory tract and the genito-urinary
tract. In some primates, they are present on the vascular
endothelium of all tissues and in chimpanzee, gorilla and
man they are additionally present on erythrocytes, hence
their name blood group antigens [2].
The molecular genetic basis of the human ABO alleles
has been elucidated. Although many mutations have been
described to date, only some of these are functionally
relevant [3]. Functional analyses have been performed to
determine which amino-acids are responsible for the A or
B enzyme activities. These studies revealed that the amino-
acids at positions 266 and 268 and to a lesser extent at
position 235 were critical in determining whether the
enzyme transfers an N-acetylgalactosamine, a galactose or
both [4]. For example, the presence of a glycine at position
268 allows the transfer of an N-acetylgalactosamine
(GalNAc). But this activity is modulated by amino-acids
at the other two positions as, depending on these, transfer
of Gal may become possible in addition to the transfer of
GalNAc.
The biological meaning of the ABO phenotypes is still
largely obscure. Yet, the above mentioned tissue distribu-
tion and some associations between the ABO polymor-
phism and infectious diseases suggest a role in the
interaction with pathogens. For example, a strong associ-
ation is found between blood group O and susceptibility to
cholera [5]. Moreover, various strains of bacteria can adhere
to either A, B or H antigens, suggesting that microbes use
them as receptors and that their host range may be
influenced by the individual blood group phenotype [6].
Histo-blood group antigens or structurally related carbo-
hydrates may be present on pathogens. Protective antibod-
ies directed against these structures may therefore be
Correspondence to J. Le Pendu, Inserm U419,
Institut de Biologie, 9 Quai Moncousu, F-44093, Nantes, France.
Fax: + 33 240 08 40 82, Tel.: + 33 240 08 40 99,
E-mail: jlependu@nantes.inserm.fr
Abbreviations: A transferase, UDP-GalNAc:Fuca1,2GalaGalNAc
transferase; B transferase, UDP-Gal:Fuca1,2GalaGal transferase;
CHO, Chinese hamster ovary; GalNAc, N-acetylgalactosamine;
Gal, galactose; Fuc, fucose; FITC, fluorescein isothiocyanate;
AEC, 3-amino-9-ethylcarbazol.
Enzymes: UDP-GalNAc:Fuca1,2GalaGalNAc transferase
(EC 2.4.1.40).
(Received 22 March 2002, revised 12 June 2002, accepted 5 July 2002)
Eur. J. Biochem. 269, 4040–4047 (2002) FEBS 2002 doi:10.1046/j.1432-1033.2002.03094.x

differentially generated by the host, depending on the blood
group phenotype [7]. The differential host range for
adhesion of pathogens and the presence of protective
antibodies mean that all individuals would not be equally
sensitive to a given pathogen. This would provide a
mechanism of protection against the pathogenic strain at
the level of the population and a selective force to maintain
polymorphism at the ABO locus [6].
In the present work, we report the isolation of a BDIX rat
cDNA homologous to the human ABO sequences encoding
for an A histo-blood group enzyme. Examination of the
tissue distribution of the corresponding mRNA and of the
A and B antigens in the BDIX strain of rat was performed,
allowing some aspects of the ABO genetics in this species to
be considered.
MATERIALS AND METHODS
Cloning of a rat ABO-like cDNA
An EMBL-3 rat genomic DNA phage library (Clontech)
was screened with a cDNA probe (P718), derived from the
human blood group A transferase and corresponding to
nucleotides 148–865 of the human cDNA sequence [8].
Positive recombinants were purified, digested with EcoRI
and analyzed by Southern Blot using the pb718 probe. Two
hybridization-positive fragments (4.0 and 3.5 kb) were
obtained and digested by HinfI. The products were ligated
into the pUC18 vector (Pharmacia) and sequenced. One
clone, from the 4 kb fragment digest, contained an insert of
406 bp, a stretch of which was 84.4% similar to exon 6 of
the human blood group A gene coding sequence. The
complete coding sequence was obtained by RACE/PCR
using primers deduced from this fragment. To this end, total
RNA from BDIX rat stomach was extracted using the SV
Total RNA isolation kit from Promega. This RNA
preparation was used to obtain mRNA using the Oligotex
kit from Qiagen. Double stranded cDNA synthesis, adaptor
ligation and RACE/PCR were performed using the Clon-
tech Marathon cDNA Amplification kit. Elongation in the
5¢direction was performed using an inverted primer
deduced from the 406 bp fragment homologous to the
human A gene (GTCGATGTTGAAGGTCCCCTCCCA
GATG) and the adaptor AP1 primer provided by the
supplier. The second nested PCR was performed using a
nested inverted primer deduced from the same fragment
sequence (TCCCAGATGATGGGAGCCACGCCAA
GG) and the nested adaptor AP2 primer from the supplier.
Synthesis in the 3¢direction was performed by the same
method using the following first primer
(CTTGTCTTCACTCCTTGGCTGGCTCCCAT) and
the adaptor AP1 primer, followed by a second nested
PCR with the second nested primer (CATCTGG
GAGGGGACCTTCAACATCGAC). The PCR was run
with the Advantage Polymerase (Clontech) and the man-
ufacturer’s touchdown-RACE program. The PCR products
were ligated into the pUC18 vector and sequenced. To
obtain the full coding fragment, stomach cDNA was PCR
amplified using the following primers, deduced from the
sequence of the 5¢and 3¢RACE products: AC
CATCCCGGGCCTTGCATGGA (forward) and GCTA
CAGGTACCGCCTCTCCAA (reverse). The product was
ligated into the pUC18 vector and sequenced.
Sequence analysis
Multiple alignments were performed with the
CLUSTALW
program [9]. Genetic distances of the complete deduced
peptide sequences were calculated by the Neighbor joining
method from the
PHILIP
program. Approximate location of
transmembrane regions were determined using the
TMHMM
,
TMAP
and
TOPPRED
2 programs. Determination of the exon/
intron boundaries were obtained by analysis of the rat
genomic sequences available in the NCBI database. The
programs used are all available from http://www.infobio-
gen.fr.
Chromosome localization
The Abo gene was first assigned to a rat chromosome using
a panel of standard rat X mouse cell hybrids that segregate
rat chromosomes [10]. The hybrids were typed by PCR
with the following primers: 5¢-GGAGCAGCTGGAGT
CATG-3¢and 5¢-GGTCATCCTGTATCCTTCA-3¢(the 5¢
end of these primers corresponds to the positions 163 and
270, respectively, of [35104745] from the Rattus norvegicus
WGS trace database). For regional localization, the panel of
rat ·hamster radiation cell hybrids [11] was typed in the
same manner. The mapping results were obtained from the
rat radiation hybrid map server at the Otsuka GEN
Research Institute (http://ratmap.ims.u-tokyo.ac.jp/menu/
RH.html) [11].
RT-PCR analysis
Total RNAs (1 lg) from various rat tissues listed below
were prepared using the SV Total RNA isolation System kit
from Promega and reverse transcribed at 42 Cwiththe
M-MLV reverse transcriptase from Promega. Contaminat-
ing DNA had been removed by digestion with RNAse-free
DNAseI (10 unitsÆlg
)1
RNA) for 15 min at room temper-
ature. Amplification of the cDNA corresponding to the A
enzyme cDNA was performed with the following primers:
CAGACGGATGTCCAGAAAGTTG and: GCTACAG
GTACCGCCTCTCCAA. Amplification was performed
using the Advantage Polymerase (Clontech) with initial
denaturation at 94 C 3 min, followed by 30 cycles of 94 C
30 s, 64 C45s,68C 2 min. The amplification yields a
product of 929 bp. The control of cDNA quality was
performed by amplification of glyceraldehyde phosphate
dehydrogenase (GAPDH).
Transfection of CHO cells
The complete coding sequence of the rat A gene was
inserted into the pDR2 eukaryotic expression vector
(Clontech) deleted of the sequences lying between the
EcoRV and ClaI sites. Chinese hamster ovary carcinoma
cells, CHO cells, are devoid of a1,2fucosyltransferase
activity and therefore of ABH antigens. They were first
transfected using LipofectAMINTM with the rat a1,2fuco-
syltransferases cDNA FTB (GenBank accession number
AF131238) in the pBK-CMV expression vector (Gibco,
Paisley, UK) according to the manufacturer’s instructions.
Twenty-four hours later, fresh medium was added and 48 h
later, selective medium containing 0.6 mgÆmL
)1
G418
(Gibco) was added. Transfected cells expressing a1,2-linked
FEBS 2002 Rat histo-blood group A enzyme (Eur. J. Biochem. 269) 4041
fucose residues were selected by flow cytometry, using
fluorescein isothiocyanate (FITC)-labeled UEA-I, after
cloning by limiting dilutions as previously described [12].
For each cell line, a strongly expressing clone was selected
and transfected a second time using the same procedure
with the rat A enzyme cDNA in the pDR2 vector. Forty-
eight hours later, cells were cultured in selective medium
containing 0.6 mgÆmL
)1
hygromycin. After cloning by
limiting dilutions, strongly A antigen expressing transfec-
tants were selected. Control transfected cells were prepared
by transfection with the empty vectors. These stable
transfectants were cultured in RPMI 1640, 10% fetal
bovine serum, 2 m
ML
-glutamine, free nucleotides
(10 lgÆmL
)1
), 100 UÆmL
)1
penicillin and 100 lgÆmL
)1
streptomycin (Gibco) supplemented with 0.25 mgÆmL
)1
G418 and 0.2 mgÆmL
)1
hygromycin. They were cultured at
confluence after dispersal with 0.025% trypsin in 0.02%
EDTA. Cells were routinely checked for mycoplasma
contamination by Hoescht 33258 (Sigma, St Louis, MO,
USA) labeling.
Cytofluorimetric analysis
Viable cells (2 ·10
5
per well) were incubated with antibod-
ies at the appropriate dilutions in NaCl/P
i
containing 0.1%
gelatin for 1 h at 4 C. Optimal concentrations of antibodies
were chosen after serial dilutions to obtain the strongest
positive signal without cell death. The anti-A mAb 3–3 A
was obtained from J. Bara (INSERM U482, Villejuif,
France). It recognizes all types of A antigens [13] and does
not show any detectable cross reactivity with B epitopes as
judged from enzyme immunoassay with synthetic oligosac-
charides and immunostaining of human tissues of known
ABO phenotypes. The anti-B mAb ED3 is a gift from
A. Martin (CRTS, Rennes, France). It recognizes all types
of B and shows no detectable cross-reactivity with
A epitopes or with the Gala1,3Gal epitope [14]. Following
the first incubation with monoclonal antibodies, after three
washes with the same buffer, a second 30 min incubation
was performed with the FITC-labeled anti-(mouse Ig) Ig
under the same conditions. After washings in the same
buffer, fluorescence analysis was performed on a FACScan
(Beckton–Dickinson) using the
CELLQUEST
program.
Detection of enzyme activity
Confluent transfected CHO cells were rinsed with ice-cold
NaCl/P
i
, pH 7.2, then recovered by scraping. After
washing with ice-cold NaCl/P
i
, cells were solubilized in
50 m
M
cacodylate pH 7.0, containing 2% (v/v) Triton
X-100 on ice for 30 min Following a centrifugation at
13 000 gfor 10 min, the supernatant was collected and used
as a crude enzyme preparation. Protein concentration was
determined using bicinchoninic acid. The reaction mixture
contained: 50 lg protein extracts, 30 m
M
MnCl
2
,5m
M
ATP, 10 m
M
NaN
3
,5m
M
2¢fucosyllactose and 20 l
M
UDP-
D
-[
14
C]N-acetylgalactosamine (55 mCiÆmmol
)1
,ICN,
Costa Mesa, CA, USA) or 20 l
M
UDP-
D
-[
14
C]galactose
(278 mCiÆmmol
)1
, NEN, Chemical Center, Dreieichen-
dain, Germany) in a final volume of 50 lLandwas
incubated at 37 C for 16 h. After incubation, the reaction
mixture was quenched with 750 lL distilled water and
applied to an AG1-X8 column, chloride form, 100–200
mesh (Bio-Rad, Hercules, CA, USA). The radiolabeled
product was then eluted with 1 mL water and counted in
5 mL scintillation liquid (Ready SafeTM, Beckman, Palo
Alto, CA, USA). Background levels of radioactivity were
obtained from controls without exogenous acceptor.
Values obtained for the controls were then subtracted
from those obtained for the assays.
Immunohistological analysis
Tissues from 2- to 3-month old rats were collected and
immediately frozen or paraffin embedded. Sections (5 lm)
werepreparedandwashedinNaCl/P
i
. Endogenous
peroxidase was inhibited using methanol/H
2
O
2
0.3% for
20 min. Sections were then washed in NaCl/P
i
for 5 min
and covered with NaCl/P
i
/BSA 1% for 20 min at room
temperature in a moist chamber. After washing in NaCl/P
i
,
sections were covered with either the primary antibodies
diluted in NaCl/P
i
/BSA 1% and left at 4 Covernight.
Sections were then rinsed thrice with NaCl/P
i
and incubated
with biotinylated anti-(mouse IgG) Ig (Vector Laboratories,
Burlingame, CA, USA) diluted at 1/100 for 60 min at room
temperature. After washing in NaCl/P
i
, the sections were
covered with peroxidase-conjugated avidin (Vector Labo-
ratories) diluted at 1 : 1000 for 45 min, washed with NaCl/
P
i
and reactions were revealed with 3-amino-9-ethylcarbazol
(AEC). Counterstaining was performed with Mayer’s
hemalun.
RESULTS
Sequence analysis
A coding sequence homologous to the human ABO coding
sequences was isolated (GenBank accession AF264018). It
possesses an open reading frame of 1047 bp, 77% identical
with the human A gene coding region. Comparisons of this
sequence with genomic sequences available in the rat
genome data bank allowed determination of the exon/
intron boundaries as they conform to the GT-AG consensus
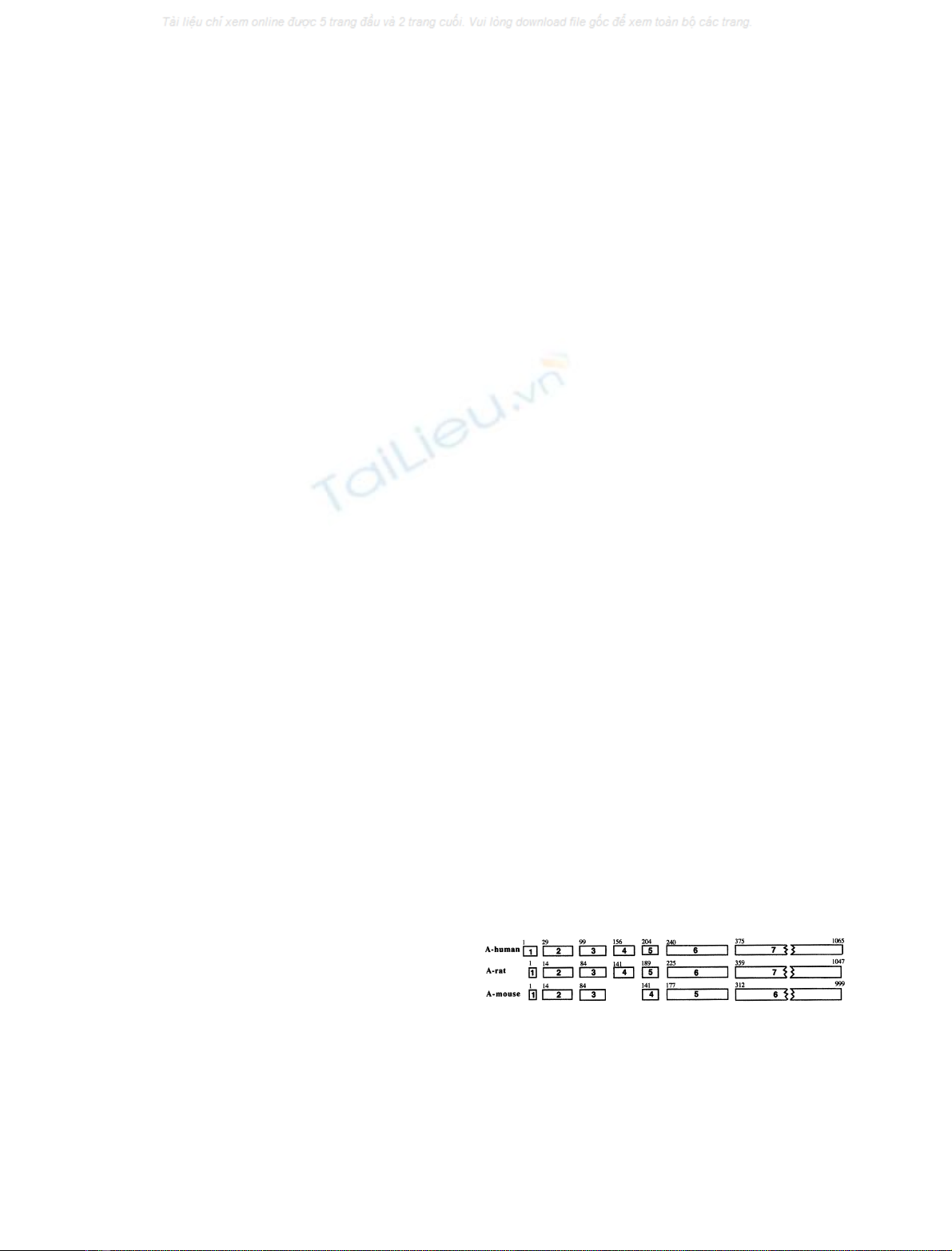
rule (Fig. 1). The gene organization appears similar to that
of the human ABO gene with seven exons [15]. The same
analysis was performed on the mouse A/B gene orthologous
to the human ABO gene [16]. This gene lacks exon 4. At the
amino-acid level, the rat sequence shows 70, 71 and 77%
identity with the human, pig and mouse sequences, respec-
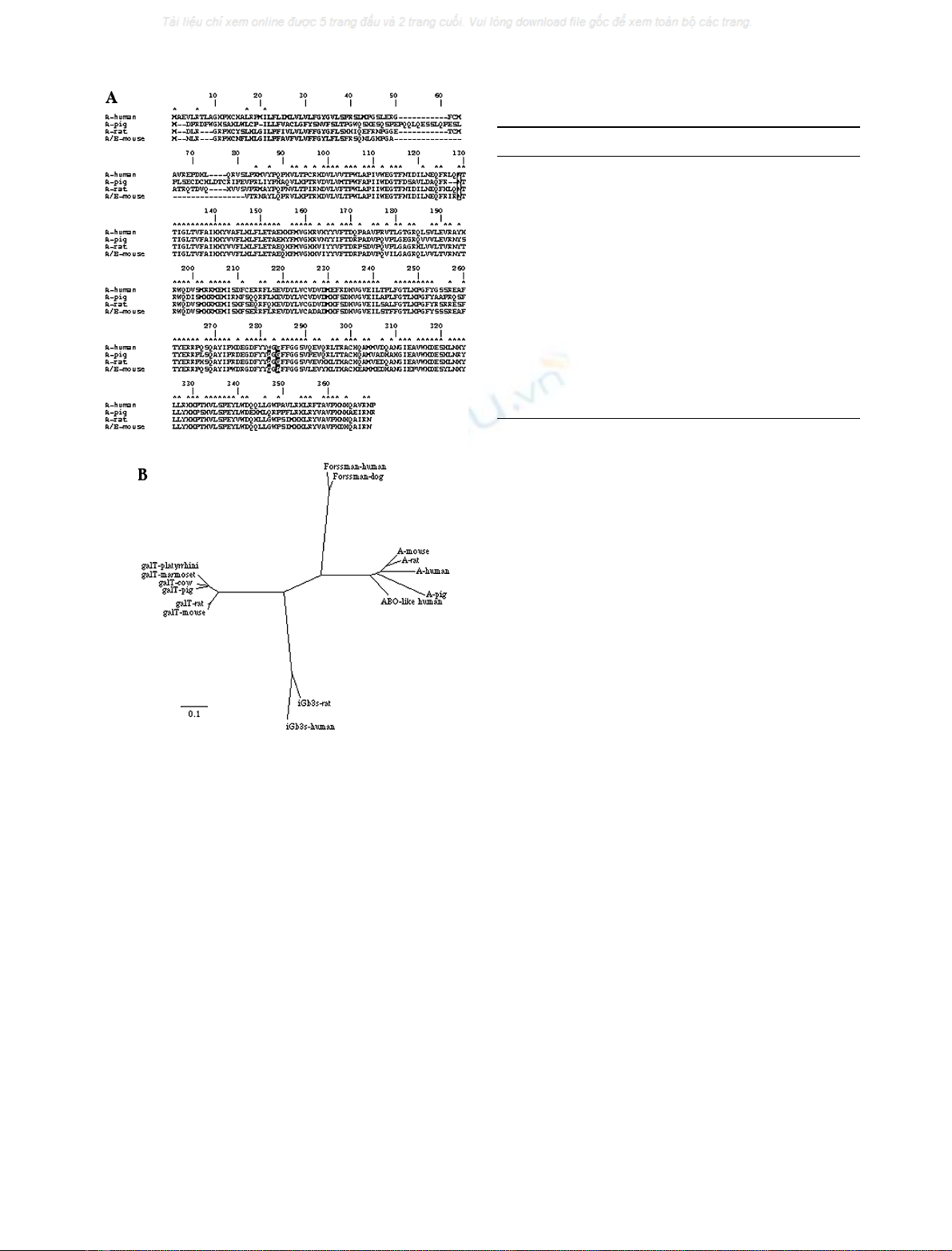
tively (Fig. 2A, Table 1). Like all glycosyltransferases the
Fig. 1. Comparison of the rat Abo gene exonic structure with those of
the human and mouse orthologs. Organization of the human and mouse
genes has been previously reported. Exons are represented by boxes
numbered in bold. Nucleotide numbers limiting the exons are noted in
superscript. For the mouse gene, a search in the mouse genomic data
bank confirmed the absence of one exon, corresponding to exon 4 of
the other two species, as the genomic sequence separating mouse exons
3 and 4 revealed no potential exonic sequence with homology to the
human and rat exon 4.
4042 A. Cailleau-Thomas et al. (Eur. J. Biochem. 269)FEBS 2002
rat enzyme presents a short N-terminal intracytoplasmic
domain, a transmembrane domain followed by a stem
region and a catalytic domain, the latter domain presenting
the highest identity among the four sequences. Of note, the
presence of a conserved potential N-glycosylation site at
the beginning of the catalytic domain. Previous studies of
the human ABO transferases underscored the major
importance of the amino-acid at position 268 with a glycine
determining A activity whereas an alanine determines B
activity. A glycine is present at the homologous position in
the rat sequence (position 263), suggesting a potential A
enzyme activity.
At present, the ABO gene family is known to comprise
four members, the ABO gene itself, the a3galactosyltrans-
ferase (pseudo B) gene [17], the aN-acetylgalactosaminyl-
transferase or Forssman synthetase gene [18] and the
a-galactosyltransferase iGb3 synthetase gene [19]. Phylo-
genetic analysis of this gene family revealed a clear
distinction between the four genes, with the new rat
sequence falling within the ABO cluster (Fig. 2B).
Chromosome localization of the rat Abo gene
The gene was first assigned to rat chromosome 3, using a
panel of 16 standard rat X mouse cell hybrid clones
segregating rat chromosomes. No discordant clone was
obtained for chromosome 3, while at least two discordant
clones were counted for each other chromosome (data not
shown). Chromosome placement by radiation hybrid map-
ping confirmed this result, with a precise localization
between D3Rat54 (at 58cR, lod score ¼5.67) and
D3Rat50 (at 62cR, lod score ¼5.12). This position corre-
sponds to the centromeric region of the chromosome (bands
3q11–q12). This rat chromosome region is known to be
homologous to the human region 9q34 [20,21], where the
human ABO gene resides [22].
Determination of the enzyme activity
In order to study the functional characteristics of the rat
A-like gene, the cDNA was transfected in CHO cells already
stably transfected with an a2fucosyltransferase cDNA,
namely the rat FTB [23]. The presence of this fucosyltrans-
ferase in CHO cells allows expression of H histo-blood
group structures which are compulsory precursors of the A
and B antigens. Stable doubly transfected cells were isolated
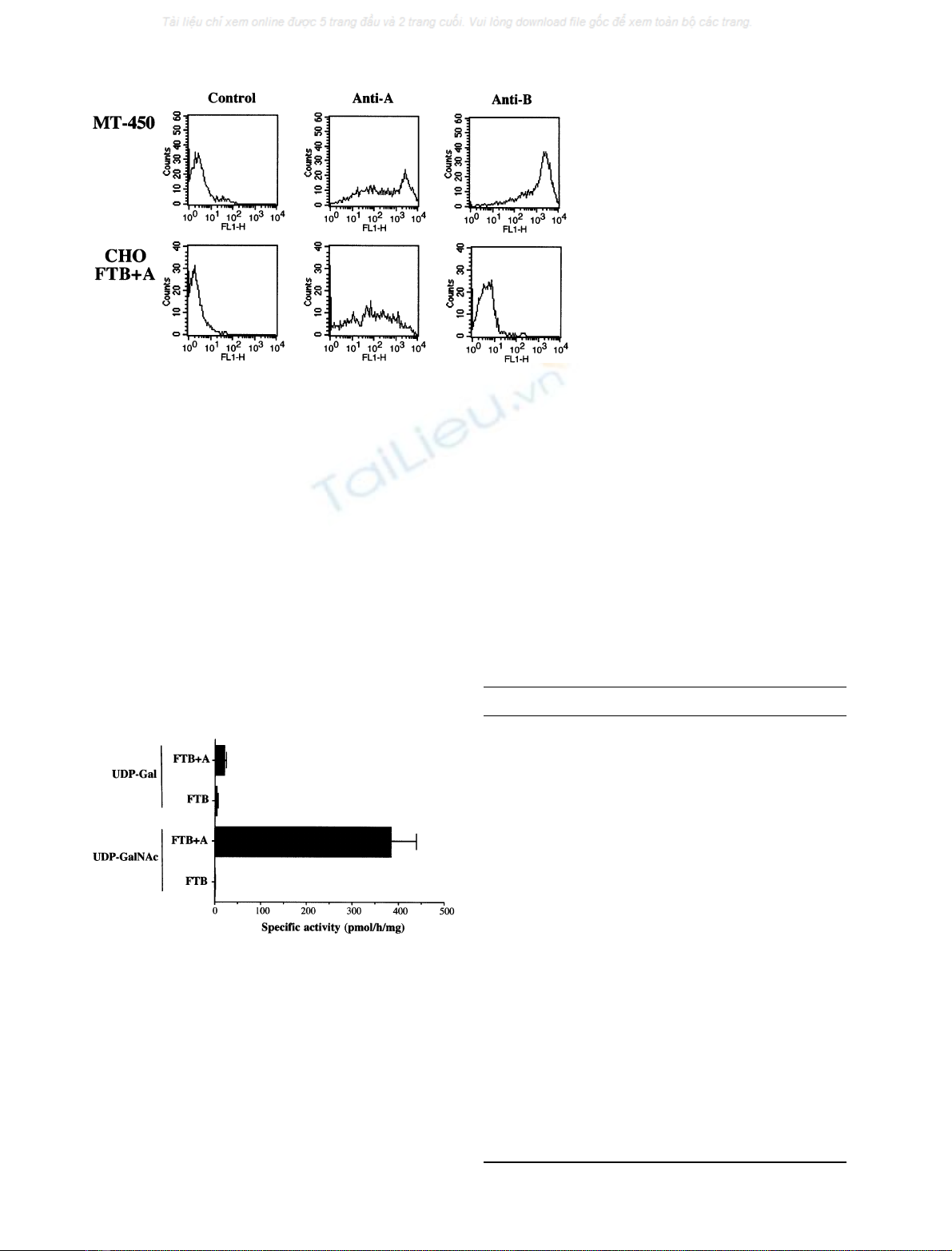
and tested by flow cytometry for their expression of A or B
antigens (Fig. 3). A positive control cell line (MT-450)
known to constitutively express both antigens indicated that
the antibodies readily detected their respective epitopes
when present [24]. The doubly transfected CHO cells were
strongly labeled by the anti-A reagent, but not significantly
Fig. 2. Comparison of the amino acid sequences of the A or cis A/B
transferases in four species (A) and phylogenetic analysis of the ABO
gene family (B). (A) Identical residues are marked by arrows. The
transmembrane regions are highlighted in grey, the conserved N-gly-
cosylation site is boxed and the amino-acids corresponding to positions
266 and 268 of the human sequence are labeled in white on black
boxes. The numbering corresponds to that of a consensus sequence.
(B) Genetic distances were calculated from
CLUSTALW
multiple align-
ments using the neighbour joiningmethod from the sequences listed
in Table 1. The scale bar represents the number of substitutions per site
for a unit branch length.
Table 1. Identification of the ABO gene family sequences used for the
phylogenetic analysis.
Enzyme Species GenBank/EBI
A transferase Human J05175
A transferase Rat AF264018
A transferase Pig AF050177
A(cis A/B) transferase Mouse AB041039
A-like
a
Human M65082
Gal transferase Platyrrhini S71333
Gal transferase Marmoset A56480
Gal transferase Cow J04989
Gal transferase Pig L36152
Gal transferase Mouse M85153
Gal transferase Rat AF520589
IGb3 synthetase Human AL513327
IGb3 synthetase Rat AF246543
Forssman synthetase
a
Human AF163572
Forssman synthetase Dog CFU66140
a
Pseudogenes.
FEBS 2002 Rat histo-blood group A enzyme (Eur. J. Biochem. 269) 4043
by the anti-B reagent, suggesting that the new rat gene
encodes an enzyme with the catalytic activity of the A histo-
blood group transferase. To confirm this result, the enzyme
activity was directly assayed on cell extracts of CHO
transfectants (Fig. 4). No transfer of galactose or
N-acetylgalactosamine could be detected on extracts from
the control FTB transfected cells using 2¢fucosyllactose as
acceptor. However, cell extracts from the double transfec-
tants showed a high N-acetylgalactosaminyltransferase
activity and a weak galactosyltransferase activity. These
results indicate that the new rat enzyme is indeed an A histo-
blood group transferase with a small B transferase activity.
Tissue expression of the mRNA and of the A
and B antigens in the rat
An RT-PCR analysis was performed to determine the
tissue expression of the rat Abo gene. Primers were chosen
to encompass different exons so as to confirm amplifica-
tion of cDNA and lack of contamination by genomic
DNA. A band at the expected size was detected in various
tissues as indicated in Table 2. A strong signal was
obtained in the oesophagus, the stomach, the colon, the
Fig. 3. Cytofluorimetric analysis of cell surface
A and B antigens of stably transfected CHO
cells. CHO cells previously transfected with
the rat FTB cDNA encoding an a1,2fucosyl-
transferase were cotransfected with the rat A
enzyme cDNA. Stable transfectants were iso-
lated and one of these clones was used for
analysis. The MT-450 cell line, a mammary
carcinoma cell line from the w/Fu rat strain,
was used as positive control. The A and B
antigens were detected using the 3–3 A and
ED3 Mabs, respectively. The Logs of fluo-
rescence intensities are plotted against cell
numbers. Negative controls were performed in
absence of primary antibody.
Fig. 4. Enzymatic assays of CHO transfectant cell extracts. The same
stable transfectants of CHO cells used for the cytofluorimetric analysis
of the A or B antigens expression (Fig. 3) were used to assay the
enzyme activity. Cell extracts were prepared as described in the
Materials and methods section. Activities were determined using
2¢fucosyllactose as acceptor substrate and either UDP-[
14
C]galactose
or UDP-[
14
C]N-acetylgalactosamine as donor substrates. The product
of the reaction was separated on AG1-X8 anion exchange columns.
The background was determined in absence of acceptor substrate and
its value was deduced from the values obtained in the presence of the
acceptor. Values of the specific activities are given in
pmolÆh
)1
Æmg protein
)1
of either [
14
C]galactose or [
14
C]N-acetylgalac-
tosamine transferred.
Table 2. Tissue distribution of the Abo mRNA and of the A and B
antigens in the BDIX rat. Transcripts were detected by RT-PCR from
total RNA extracts of various rat tissues using specific primers. An
indication of the intensity of the detected band is given, with +++
corresponding to the strongest signal and – to no detectable signal. The
A and B antigens were detected by immunohistochemistry on frozen
tissue sections using well characterized specific monoclonal antibodies.
In tissues positive for both A and B antigens, the cellular distribution
of the two antigens is not always identical (see text for details). The
labeling by the anti-A antibody was the same on paraffin embedded
sections, but no B antigen was detected on such sections. ND ¼not
done.
Tissue mRNA A antigen B antigen
Tongue ND +++ –
Oesophagus ++ ++ –
Stomach ++ ++ ++
Small intestine – – –
Caecum ND +++ +++
Large intestine +++ +++ ++
Pancreas + +++ ++
Parotid gland ND +++ –
Submaxillary gland +– ++ ++
Liver – – –
Trachea ND – –
Lung – – –
Kidney ++ – ++
Urinary bladder ++ – –
Ovary – – –
Uterus ++ +++ –
Testis – – –
Seminal vesicle ND +++ –
Thyroid gland ND +++ –
Parathyroid gland ND +++ –
Brain – – –
Muscle +– – –
Skin – – –
Spleen +– – –
Thymus ++ ++ –
Lymph node – – –
4044 A. Cailleau-Thomas et al. (Eur. J. Biochem. 269)FEBS 2002


























