
REVIEW ARTICLE
Structures of human proteinase 3 and neutrophil
elastase – so similar yet so different
Eric Hajjar
1
, Torben Broemstrup
2,3
, Chahrazade Kantari
4
,Ve
´ronique Witko-Sarsat
4,5
and Nathalie Reuter
3,6
1 Dipartimento di Fisica, University of Cagliari (CA), Italy
2 Department of Informatics, University of Bergen, Norway
3 Computational Biology Unit, BCCS, University of Bergen, Norway
4 Inserm, U845 and U1016, Paris, France
5 Institut Cochin, Universite
´Paris Descartes, CNRS (UMR 8104), France
6 Department of Molecular Biology, University of Bergen, Norway
Introduction
Human neutrophil elastase (hNE), human cathepsin G
(hCatG) and human proteinase 3 (hPR3) (also termed
myeloblastin [1] and p29b [2]) are serine proteases
mostly expressed in polymorphonuclear neutrophils,
but also are found in monocytes. All three enzymes
are homologous, although hNE and hPR3 share 56%
Keywords
human neutrophil elastase; inflammation;
myeloblastin; neutrophil, proteinase 3;
vasculitis; Wegener granulomatosis
Correspondence
N. Reuter, Department of Molecular
Biology, University of Bergen,
Thormohlensgt 55, N-5008 Bergen, Norway
Fax: +47 555 84295
Tel: +47 555 84040
E-mail: nathalie.reuter@mbi.uib.no
(Received 22 January 2010, revised 11
March 2010, accepted 18 March 2010)
doi:10.1111/j.1742-4658.2010.07659.x
Proteinase 3 and neutrophil elastase are serine proteinases of the polymor-
phonuclear neutrophils, which are considered to have both similar localiza-
tion and ligand specificity because of their high sequence similarity.
However, recent studies indicate that they might have different and yet
complementary physiologic roles. Specifically, proteinase 3 has intracellular
specific protein substrates resulting in its involvement in the regulation of
intracellular functions such as proliferation or apoptosis. It behaves as a
peripheral membrane protein and its membrane expression is a risk factor
in chronic inflammatory diseases. Moreover, in contrast to human neutro-
phil elastase, proteinase 3 is the preferred target antigen in Wegener’s gran-
ulomatosis, a particular type of vasculitis. We review the structural basis
for the different ligand specificities and membrane binding mechanisms of
both enzymes, as well as the putative anti-neutrophil cytoplasm autoanti-
body epitopes on human neutrophil elastase 3. We also address the differ-
ences existing between murine and human enzymes, and their consequences
with respect to the development of animal models for the study of human
proteinase 3-related pathologies. By integrating the functional and the
structural data, we assemble many pieces of a complicated puzzle to pro-
vide a new perspective on the structure–function relationship of human
proteinase 3 and its interaction with membrane, partner proteins or cleav-
able substrates. Hence, precise and meticulous structural studies are essen-
tial tools for the rational design of specific proteinase 3 substrates or
competitive ligands that modulate its activities.
Abbreviations
a1-PI, a1-proteinase inhibitor; ANCA, anti-neutrophil cytoplasm autoantibody; hNE, human neutrophil elastase; hPR3, human proteinase 3;
human cathepsin G, hCatG; mbPR3, membrane hPR3; PR3, proteinase 3.
2238 FEBS Journal 277 (2010) 2238–2254 ª2010 The Authors Journal compilation ª2010 FEBS

sequence identity for the mature enzymes, whereas
their similarity to hCatG is approximately 35%. Neu-
trophil serine proteinases are considered to be crucial
elements in neutrophil effector mechanisms [3,4]. Using
knockout mice invalidated for either hNE or hCatG, it
has been clearly demonstrated that both enzymes are
required for complete and adequate microbicidal activ-
ity [5,6]. Despite the lack of hPR3 knockout mice, a
similar function has been assigned for PR3 [2]. In
addition, it is now clear that these enzymes are also
involved in non-infectious inflammatory processes and
cell signaling [7,8]. A salient feature of hPR3 is its
identification as the main target antigen of the anti-
neutrophil cytoplasm autoantibodies (ANCA) in a
particular type of vasculitis, the Wegener’s granuloma-
tosis, which is a systemic inflammatory disease involv-
ing the lung, skin and kidney. The mechanisms
underlying this specific autoimmunization against
hPR3, and not against its homologs such as hNE, are
still unknown [9]. Because of the high sequence similar-
ity between hNE and hPR3, the substrate specificity
and the resulting functions of hPR3 have often been
extrapolated from the available data on hNE.
Together with the lack of structural and biophysical
studies on hPR3, relative to hNE, this has contributed
to a functional annotation of hPR3 that is too simplis-
tic. In recent years, however, more attention has been
paid to the structural properties of PR3 [10–14]. A bet-
ter understanding of the structure–function relation-
ship of the enzyme will contribute to the elucidation of
its original functions and help with the design of spe-
cific substrates for one or the other enzyme. So far, the
identification of the presence and hence the respective
role of either hPR3 or hNE in complex biological
models (both in vitro and in vivo) is impaired by a lack
of reliable specific inhibitors.
We review the sequence and structural data available
on both enzymes and highlight their similarities and
differences. We then summarize and discuss the latest
findings on three particular aspects of hPR3: ligand
specificity, membrane binding and putative ANCA
epitopes. We also address the similarities and differ-
ences between murine and human enzymes, and their
consequences with respect to the development of
animal models for the study of human PR3-related
pathologies.
PR3 and NE are highly similar
chymotrypsin-like serine proteases
All serine proteases are named after the nucleophilic
serine in their active site. The family of serine prote-
ases comprises four distinct clans, named after
proteins representative of each clan: chymotrypsin,
subtilisin, carboxypeptidase Y and caseinolytic prote-
ase [15]. PR3 and NE are chymotrypsin-like serine
proteases. Despite the absence of any conservation of
secondary or tertiary structure elements, the four
clans of serine proteases all have the same active site
consisting of three amino acids: His, Asp and Ser.
The relative orientation of the histidine, serine and
aspartic acid is similar in all clans and results in the
formation of strong hydrogen bonds between histi-
dine and serine on the one hand, and histidine and
aspartic acid on the other hand. By convention, in
chymotrypsin-like serine proteases, the histidine,
aspartic acid and serine are numbered 57, 102 and
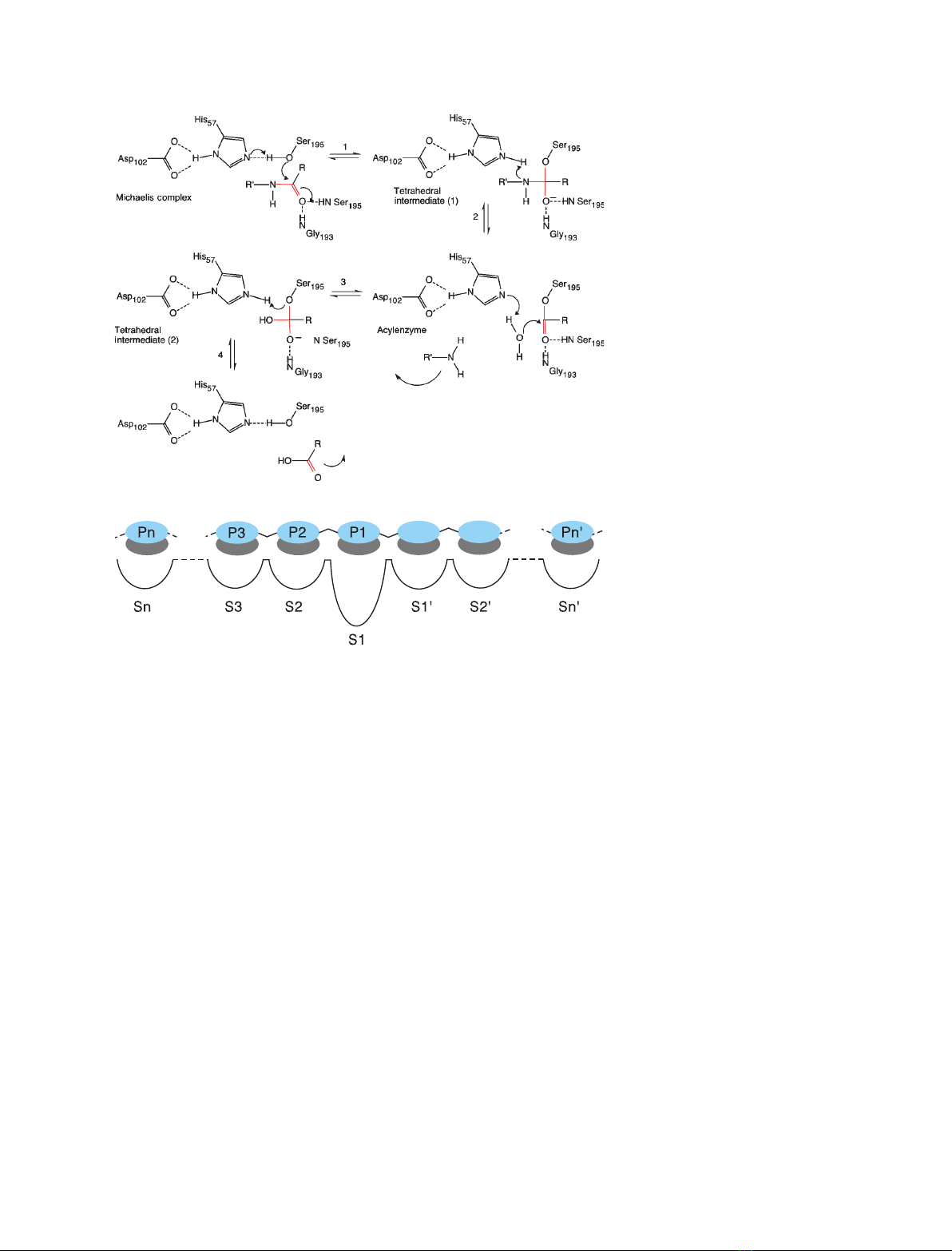
195, respectively. The reaction mechanism is illus-
trated in Fig. 1A. The substrate is positioned opti-
mally in the active site as a result of a network of
interactions that extends on both sides of the cleav-
able bonds. The interactions sites are named using
the Schechter and Berger nomenclature [16]. The rec-
ognition or subsites of the enzyme are Sn, …S1,
S1¢,…Sn¢and, for the corresponding peptide,
Pn, …P1, P1¢,…Pn¢, where P1-P1¢is the cleavable
peptide bond (Fig. 1B).
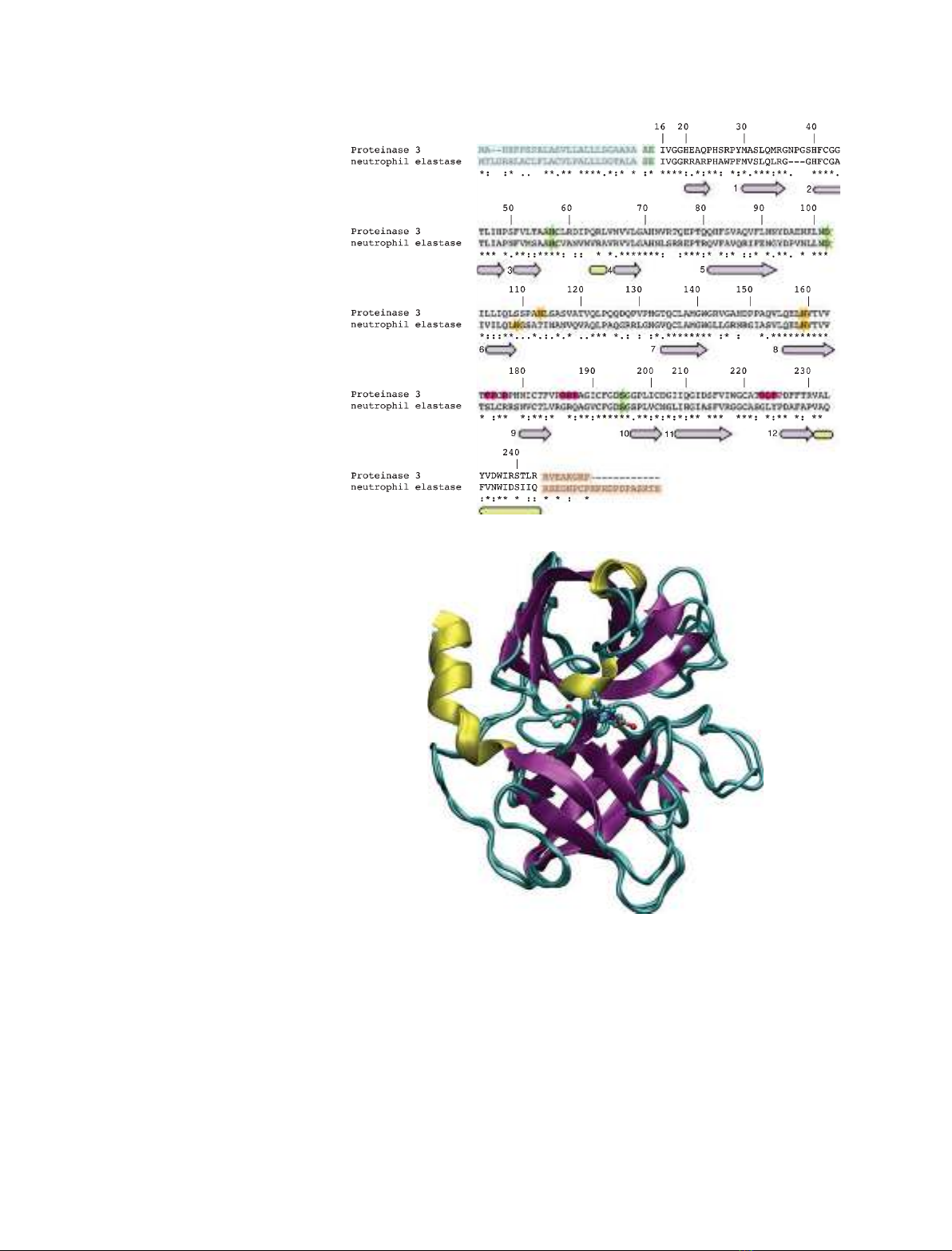
The sequences of the hNE (EC 3.4.21.37) [17–20] and
hPR3 (EC 3.4.21.76) [21–23] are shown in Fig. 2A,
where we use bovine chymotrypsinogen A numbering.
This numbering convention is used throughout the
present review (for correspondence with other number-
ing schemes, see Table S1).
hPR3 and hNE are synthesized as inactive zymogens
of 256 and 267 amino acids, respectively. These pre-
proforms undergo four consecutive steps that lead to
the mature enzymes [24–28]. The signal peptides (27
and 29 amino acids for hPR3 and hNE, respectively;
light blue boxes in Fig. 2A) are removed to yield the
proforms. The hPR3 proform is then glycosylated on
amino acids Asn113 and Asn159, and hNE on Asn
109 and Asn159 (green stars in Fig. 2A). Subse-
quently, the N-terminal dipeptide (AE for hPR3, SE
for hNE; light green boxes in Fig. 2A) is cleaved by a
cysteine protease, cathepsin C. The cleavage of the
dipeptide leads to a structural rearrangement of the
N-terminal region, which, from an extended solvent-
exposed conformation, becomes inserted into the pro-
tein core and interacts with Asp194. The enzymes then
become catalytically active. The fourth step is the
cleavage of the C-terminal pro-peptides (orange boxes
in Fig. 2A).
hPR3 and hNE are homologous and their mature
forms, comprising 221 and 218 amino acids, respec-
tively, share 56% sequence identity. Conserved resi-
dues are spread rather equally along the sequences.
E. Hajjar et al. Structure–function relationship of PR3 versus NE
FEBS Journal 277 (2010) 2238–2254 ª2010 The Authors Journal compilation ª2010 FEBS 2239
Fold and surface properties
X-ray data
Like all chymotrypsin-like serine proteases, hPR3 and
hNE adopt a fold consisting of two b-barrels made
each of six anti-parallel b-sheets (Fig. 2B). The struc-
tures of the mature forms of the human enzymes were
revealed by X-ray crystallography in the late 1980s for
hNE, and some years later for hPR3. To date, there
are seven structures of hNE deposited in the Protein
Data Bank (PDB code: 1B0F [29], 1H1B [30], 1HNE
[31], 1PPF [32], 1PPG [33], 2RG3 [34] and 2Z7F [35])
and one of hPR3 (1FUJ [36]). No structures of NE or
PR3 from other species are available (only computa-
tional models of the murine species have been
described) [11,14], nor are structures of the proforms.
hNE crystals all contain monomers (1B0F, 1HNE,
1PPF, 1PPG, 2RG3 and 2Z7F) or dimers (1H1B),
whereas hPR3 was crystallized as a tetramer, which
can be regarded as a dimer of dimers; two monomers
in a dimer are oriented so that their active sites face
each other, preventing the binding of large substrates.
Moreover, the hole in the middle of the tetramer is
lined with hydrophobic residues. This characteristic of
the crystals of hPR3 has not been observed in the case
of hNE, although it might be related to an increased
propensity of monomeric hPR3 to interact with hydro-
phobic environments through this region. Both hPR3
and hNE contain the same four disulfide bridges
between cysteine pairs 42–58, 136–201, 168–182 and
191–220.
The overall structural differences between hNE and
hPR3 are very small. The rmsd, calculated after struc-
tural alignment with stamp [37], on C-alpha atoms of
hPR3 and all seven available hNE structures is below
1A
˚and the structural difference between the seven
hNE structures is in the range 0.3–0.6 A
˚. The loop
between extended sheets 1 and 2 (amino acids 36–39)
is the only region showing a significantly larger struc-
tural variation as a result of the insertion of three resi-
dues (NPG) in hPR3 (Fig. 2A).
Different glycosylation sites
Most structures of hNE (1B0F, 1H1B, 1PPF, 1PPG
and 2Z7F) reveal the presence of two sugar moieties
on both Asn109 and Asn159, whereas one of the latest
A
B
P1′P2′
Fig. 1. (A) Reaction mechanism of serine
proteases. (1) The first step of the catalytic
reaction, after the formation of the enzyme–
substrate or Michaelis complex, is the acyla-
tion step; it starts with the attack of the
catalytic serine on the carbonyl group of the
cleavable amide bond and the transfer of
the hydroxyl hydrogen of the serine to the
histidine. (2) This leads to the release of the
C-terminal end of the substrate and the
formation of a covalent intermediate (i.e. the
acyl enzyme) between the enzyme and the
N-terminal part of the substrate. (3) The
second step of the reaction (termed deacyl-
ation) starts with the attack of a nucleophilic
water on the substrate carbonyl and
(4) ends with the release of the N-terminal
part of the substrate, when the catalytic
triad is regenerated. The nitrogen atoms of
residues Gly193 and Ser195 stabilize the
so-called oxyanion hole. (B) Schechter and
Berger convention for the numbering of
enzyme-ligand binding sites.
Structure–function relationship of PR3 versus NE E. Hajjar et al.
2240 FEBS Journal 277 (2010) 2238–2254 ª2010 The Authors Journal compilation ª2010 FEBS
structures shows only one occupied site (Asn159 for
2RG3). In the structure of hPR3, only one glycolysa-
tion site (Asn159 and not Asn113) is occupied by a
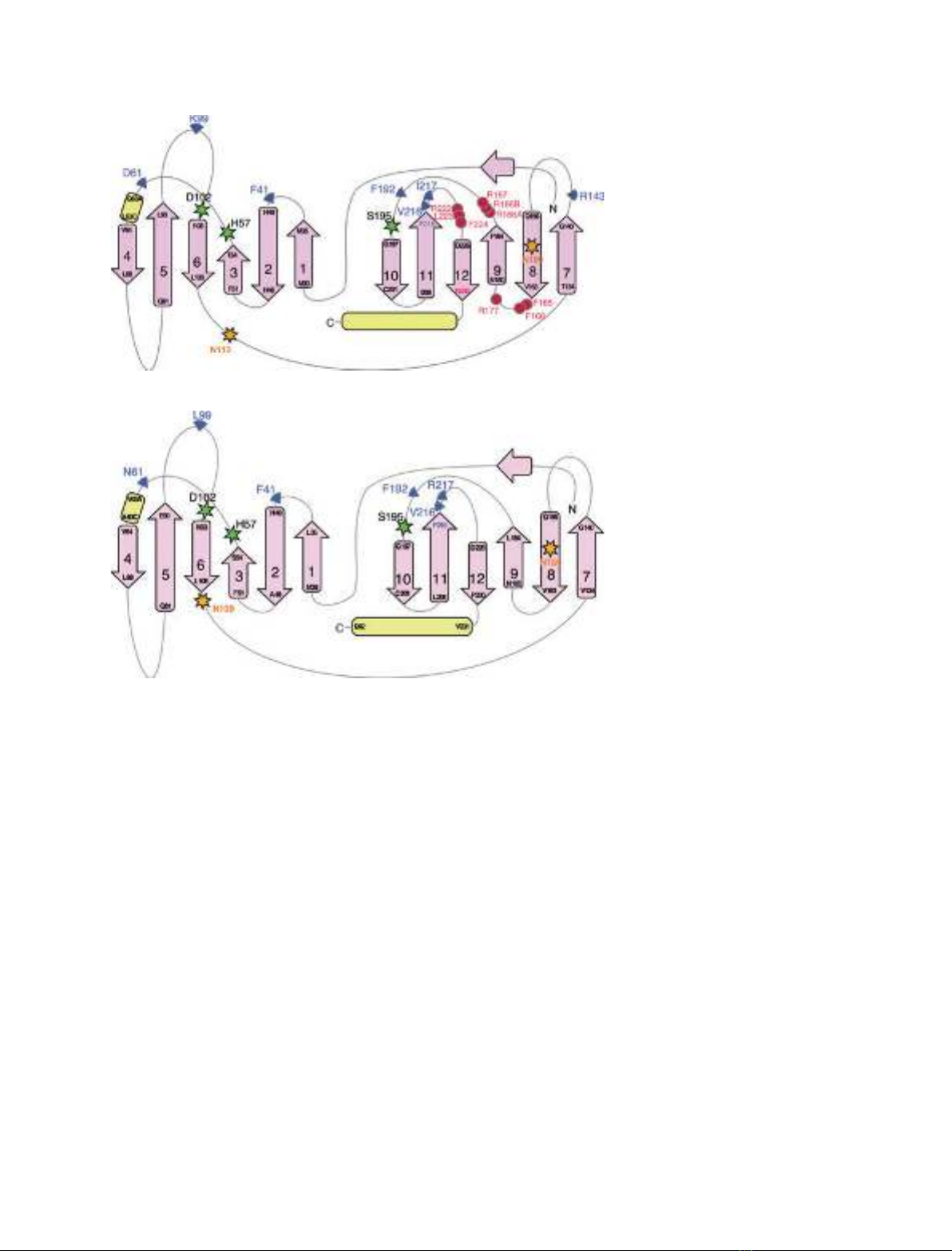
sugar. All three glycosylation sites (Asn109, Asn113
and Asn159) are remote from the catalytic triad and
the ligand binding sites (Figs 3 and 4A). According to
Specks et al. [38], both sites are occupied in neutrophil
hPR3, although they have different functional signifi-
cance; glycosylation on Asn159 influences hPR3
thermostability and increases significantly the catalytic
activity measured on a small peptidic substrate
(N-methoxysuccinyl-Ala-Ala-Pro-Val), whereas glyco-
sylation on Asn113 appears to be important (but
is not an absolute requirement) for an efficient
N-terminal processing of hPR3. Interestingly, of both
glycosylation sites, Asn113 is the furthest away from
the N-terminal end of hPR3, as observed from the
structure of the mature form. Unfortunately, no
A
B
Fig. 2. Sequence alignment and superimpo-
sition of the 3D structures of hNE and hPr3.
(A) Sequence alignment. The numbering
follows the chymotrypsin convention. Amino
acids present in the proforms are
highlighted with boxes of different colors:
signal peptides, blue; N-terminal dipeptides,
green; C-terminal propeptides, orange.
Green stars are used to highlight the amino
acids of the catalytic triad (His57, Asp102,
Ser195), whereas orange stars highlight the
glycosylation sites. The secondary structure
elements are conserved in both proteins
and are represented below the sequences
(pink arrows, extended strands; yellow
cylinders, helices). The extended strands
constituting the b-barrels are numbered 1–6
and 7–12 for the first and second barrels,
respectively. (B) Superimposition of the 3D
structures of hPr3 (1FUJ) [36] and hNE
(1PPF) [32]. Secondary structure elements
are colored as shown in Fig. 1. The catalytic
triad is represented in green balls and
sticks. Two cylinders (black lines) represent
the limits of the two b-barrels.
E. Hajjar et al. Structure–function relationship of PR3 versus NE
FEBS Journal 277 (2010) 2238–2254 ª2010 The Authors Journal compilation ª2010 FEBS 2241
structure of any of the proforms of hPR3 or hNE is
available, although X-ray structures of proforms of
homologues have been reported (pro-granzyme K,
1MZA [39], chymotrypsinogen, 2CGA [40], and tryp-
sinogen, 2TGT [41]), where it can be clearly seen that
the N-terminal end (residues 14–17) freely extends out
of the core of the protein along the extended sheet
containing residue 159 (extended sheet numbered 8
in Fig. 3). Residue 159 in chymotrypsinogen and
pro-granzyme K is thus very close to the extended
N-terminal segment.
Different surface properties
Earlier studies have questioned the relationship
between the net charge difference and the functions
of PR3 and NE [36,42]. Indeed, at neutral pH, hNE
has a net charge of +10 (it contains 19 arginines and
only nine acidic residues), whereas hPR3 has a much
lower net charge of +2, although it contains approxi-
mately the same number of charged amino acids as
hNE. The sequence of the mature form contains 13
arginines, two lysines, ten aspartic acids and four
glutamic acids. Fujinaga et al. [36], as well as
subsequent in silico pK
a
calculations [10,43], suggest
Asp213 to be protonated. The availability of the
enzymes atomistic structures allows the calculation of
the electrostatic surface potential (i.e. the electrostatic
potential created by all the amino acids of the
enzyme in its vicinity) (Fig. 5). It is a critical determi-
nant of its surface properties and it is more relevant
to its structure–activity relationship because it reflects
not only the net charge, but also the charge distribu-
tion. Electrostatic interactions are known to play a
key role in macromolecular interactions (e.g. with
partner proteins, ions or membrane binding); thus,
they might explain protease-specific functions. Inter-
estingly, in the case of hNE, there is an omnipresence
of electropositive potential that covers most of the
surface of the enzyme, except at the substrate-binding
site. This is not the case for hPR3, where electroposi-
tive clusters (or ‘patches’) alternate with negative and
A
B
Fig. 3. Topology of hPr3 (A) and hNE (B).
Pink arrows, extended strands; yellow
cylinders, helices; green stars, catalytic
triad; orange stars, glycosylation sites; pink
circles, putative membrane binding site;
blue triangles, amino acids involved directly
in ligand binding. The extended strands
constituting the b-barrels are numbered 1–6
and 7–12 for the first and second barrels,
respectively.
Structure–function relationship of PR3 versus NE E. Hajjar et al.
2242 FEBS Journal 277 (2010) 2238–2254 ª2010 The Authors Journal compilation ª2010 FEBS










![Hình ảnh học bệnh não mạch máu nhỏ: Báo cáo [Năm]](https://cdn.tailieu.vn/images/document/thumbnail/2024/20240705/sanhobien01/135x160/1985290001.jpg)















