
REVIEW ARTICLE
Long-distance interactions between enhancers and
promoters
The case of the Abd-B domain of the Drosophila bithorax complex
La
´szlo
´Sipos and Henrik Gyurkovics
Institute of Genetics, Biological Research Center, Hungarian Academy of Sciences, Szeged, Hungary
Introduction
The normal development of eukaryotic organisms
requires a precise and coordinated control of gene
expression, both spatially and temporally. In the case
of genes with a highly complex expression pattern, this
is achieved through the action of a large set of enhanc-
ers, which are often located at a considerable distance
from the regulated gene. Accordingly, one of the key
questions involved in an understanding of complex
gene regulation is how distant enhancers communicate
with their target promoters. Despite its importance,
the available scientific data relating to this question are
still extremely scarce. In this respect, one of the best-
studied systems is the regulation of the homeotic
Abdominal-B (Abd-B) gene in Drosophila.
Abd-B, one of the three genes in the bithorax complex
(BX-C), determines the identity of the posterior-most
segments in the fly. One Abd-B transcript (class A tran-
script) is responsible for the proper identity of abdom-
inal segments 5–8, while three other transcripts are
required for the identity of abdominal segment 9 and
also that of abdominal segment 10 (for examples see
[1,2]). Here we focus on the transcriptional unit coding
for the class A transcript, and refer to it and its regula-
tory regions as the Abd-B domain. The expression pat-
tern of Abd-B is regulated by a set of large (over 10 kb),
autonomous cis-regulatory domains, iab-5,iab-6,iab-7
and iab-8 in segments A5, A6, A7 and A8, respectively
(reviewed in [3,4]). As illustrated in Fig. 1A, these
cis-regulatory domains are located downstream of the
Abd-B transcription unit, and, as is the case for the other
Keywords
Abd-B; chromatin structure; Drosophila;
homeotic genes; promoter targeting
Correspondence
H. Gyurkovics, Institute of Genetics,
Biological Research Center, Hungarian
Academy of Sciences, H-6726 Szeged,
Temesvari krt. 62, Hungary
Fax: +36 62 433503
Tel: +36 62 599687
E-mail: Henrik@brc.hu
(Received 21 February 2005, accepted
10 May 2005)
doi:10.1111/j.1742-4658.2005.04757.x
Abdominal-B (Abd-B) is a complex homeotic gene with a difficult task: one
transcript determines the identity of four different abdominal segments
throughout development in Drosophila. Although an increasing amount of
information is available about the structure and the functioning of the reg-
ulatory regions that determine the expression pattern of Abd-B, it is still
not clear how these regulatory regions can contact the distantly located
(several tens of kilobases away) promoter in the nucleus, what mechanism
restricts promiscuous enhancers to this specific interaction, and how differ-
ent regulatory regions replace one another at the same promoter in subse-
quent abdominal segments. Moreover, several of these regulatory regions
have to act over chromatin domain boundaries and extensive inactive chro-
matin domains, similarly to the situation found in the chicken beta-globin
cluster. In this minireview we survey mechanisms and factors that may be
involved in mediating specific interactions between the Abd-B promoter
and its regulatory regions.
Abbreviations
Abd-B,Abdominal-B gene; BX-C, bithorax complex; Pc-G, polycomb-group; PREs, polycomb response elements; PTS, promoter targeting
sequence; trx-G, trithorax-group; TREs, trithorax response elements; tmr, transvection-mediating region.
FEBS Journal 272 (2005) 3253–3259 ª2005 FEBS 3253
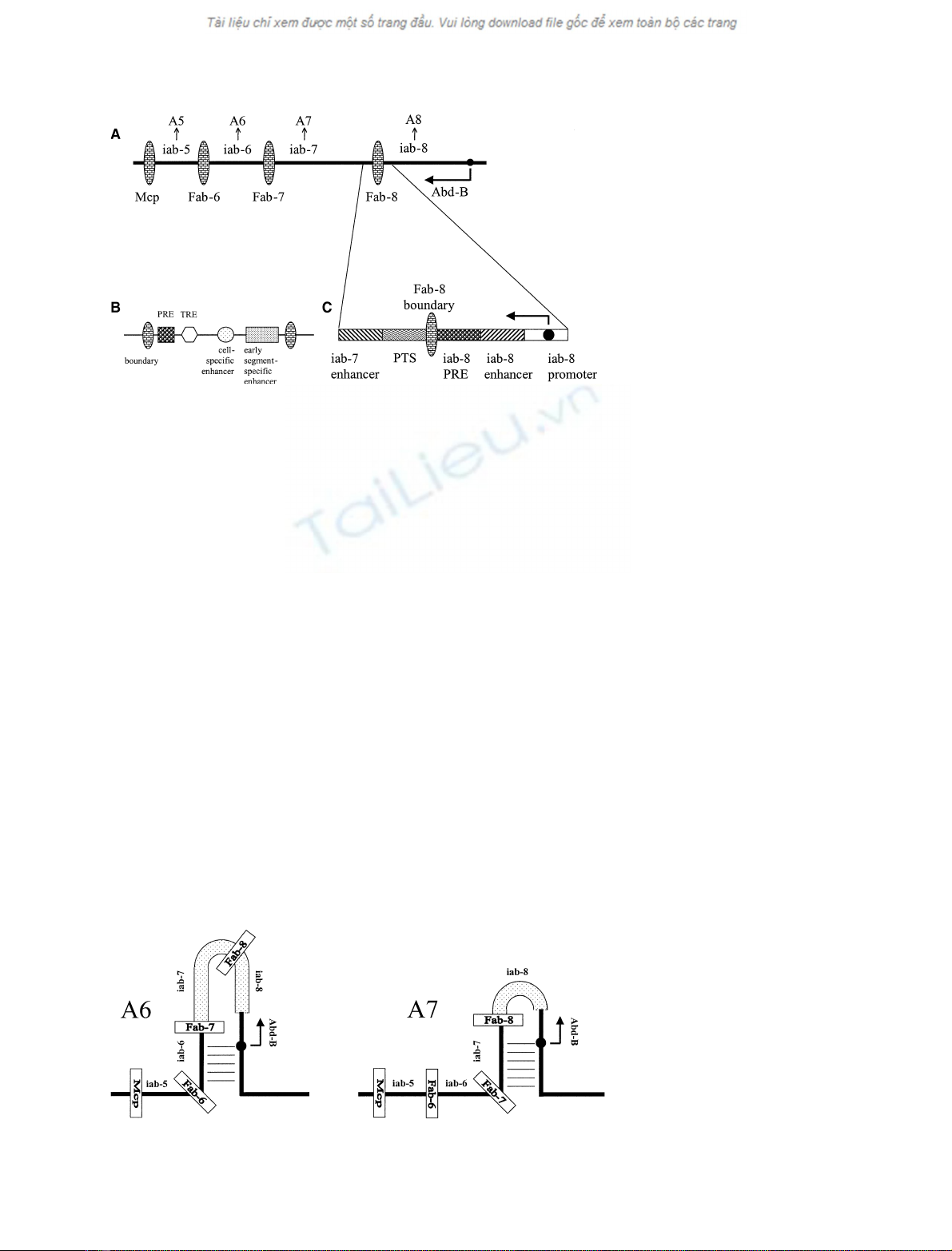
BX-C cis-regulatory domains, their proximal-distal
order along the chromosome corresponds to the anter-
ior-posterior order of the segments they specify.
Cis-regulatory regions in the Abd-B domain are
sequentially activated on proceeding from anterior to
posterior segments. In A5, for example, only one of the
four Abd-B cis-regulatory regions, iab-5, is thought to
be active, while the other three are silenced. In A6, both
iab-5 and iab-6 are active, and iab-7 and iab-8 are
silenced, but only iab-6 drives the expression of Abd-B.
Similarly, although three different Abd-B cis-regulatory
domains are active in A7, the expression of Abd-B is
directed predominantly (or exclusively) by iab-7 in this
segment of wild-type animals (Fig. 2). However, if iab-7
is deleted, the expression of Abd-B is controlled by iab-6
in both A6 and A7, resulting in the transformation of
A7 into a duplicated copy of A6, while the identity of
the more posterior segments is not altered.As expected
from this loss-of-function phenotype, the Abd-B expres-
sion pattern normally seen in A7 is replaced by an
A6-like pattern [5].
Cis-regulatory regions contain a set of different func-
tional and structural elements (Fig. 1B) identified in
transgenic reporter constructs (for example see [6]).
Among them, ‘early enhancers’ drive segmentally
restricted gene expression patterns in blastoderm
embryos as a response to the action of gap and pair-
rule gene products. Another class of enhancers iden-
tified in cis-regulatory regions are ‘cell-specific
enhancers’, which turn on reporter genes in particular
cell types without any segmental specificity. Polycomb
and trithorax response elements (PREs ⁄TREs) are
involved in generating and maintaining ‘closed’ or
‘open’ chromatin conformations, respectively, accord-
ing to the spatial activity pattern of the ‘early enhanc-
ers’. These alternative chromatin conformations will
eventually restrict the action of ‘cell-specific enhancers’
to segmental boundaries. Finally, boundary elements
flank the regulatory regions. Boundary elements can
block or greatly weaken the interactions of an enhancer
and a promoter if placed between them in transgenic
constructs, and can protect a reporter gene from the
effects of the neighboring chromatin (e.g. heterochro-
matinization) if the reporter is flanked by two of them.
The apparent function of the boundaries within BX-C
is to separate neighboring cis-regulatory regions, and to
Fig. 1. Schematic structure of the Abd-B
domain. The proximal Abd-B promoter (d)
and insulator regions (brick-patterned ovals)
separating independent 3¢cis-regulatory reg-
ions (iab-5 to iab-8) are shown (A). Each cis-
regulatory region is required for the proper
identity of one of the abdominal segments
from A5 to A8, indicated by vertical arrows.
(B) The generalized structure of a cis-regula-
tory region. (C) An enlargement of the
10 kb tmr region with the known
cis-acting elements.
Fig. 2. Model of the regulation of the Abd-B
gene in abdominal segments A6 and A7.
Although the iab-5cis-regulatory region is
also in an active conformation in A6, only
iab-6 is presumed to contact the Abd-B
promoter region (indicated by a series of
horizontal lines), while the inactive iab-7 and
iab-8 regions (thick dotted figures) loop out.
In the next abdominal segment, A7, iab-7
becomes activated and takes over the
regulation of Abd-B from iab-6.
Long-distance interactions in Abd-B L. Sipos and H. Gyurkovics
3254 FEBS Journal 272 (2005) 3253–3259 ª2005 FEBS

provide them with the autonomy necessary for inde-
pendent functioning.
The looping model
The most widely accepted model of long–range regula-
tory interactions is the looping model, which postu-
lates that enhancers and distant promoters are in
physical contact, while the intervening sequences loop
out. Although the looping model was formulated many
years ago, direct in vivo evidence for its validity has
been found only recently for the chicken beta-globin
gene cluster (reviewed in [7]). In this case, all sequences
necessary for the efficient transcription of one of the
genes in the cluster were found to be in close proxim-
ity, forming a ‘hub’, while inactive regions proved to
be pushed aside. The organization and functioning of
the Abd-B gene suggest that the looping model is also
applicable to the Abd-B regulatory unit. In A6, for
example, enhancers in iab-6 have to reach over the
entire inactive iab-7 region to act on the Abd-B pro-
moter (Fig. 2). However, the looping model raises the
question of how potentially promiscuous enhancers in
the cis-regulatory regions are able to avoid other pro-
moters, and to locate and physically approach their
proper target promoter in the viscous environment of
the nucleus.
Somatic pairing of chromosomes and
the regulation of Abd-B
A peculiarity of Drosophila is the fact that the homo-
logous chromosomes are tightly paired during the
interphase in almost all types of somatic cells (the
exceptions are cells in the early embryo), a situation
that occurs only exceptionally in most other eukaryo-
tes. Somatic pairing may affect long-distance regula-
tory interactions by interfering with loop formation. It
has been suggested that a gene may be regulated by
being switched between two states: in the case of un-
interrupted pairing of homologous sequences (‘linearly
locked state’), the enhancers are locked away from the
promoter, while in the event of local unpairing, intra-
molecular looping is allowed to promote the inter-
actions between the enhancers and the promoter [8]. In
this context, it is interesting to note that the pairing of
BX-C occurs only after the tenth hour of embryonic
development [9], eight hours later than in the case of
the histone gene cluster [10]. This difference in the tim-
ing of somatic pairing perhaps reflects the difference
between the complexities of the regulation of the two
systems: a longer time is required for the formation of
the complex looping structure in the case of BX-C,
while a shorter time is sufficient for the establishment
of the much simpler regulatory interactions of the his-
tone cluster. However, the pairing of BX-C was found
to be a dynamic process, with the paired state never
exceeding 70% of the embryonic cells at a given time
[9]. This ‘breathing’ of the paired state might be
required for the reorganization of intramolecular inter-
actions and the correction of an inappropriate looping
structure in later stages of development.
If the uninterrupted pairing of homologs is consid-
ered to be an obstacle to loop formation, then there is
an intrinsic interest in well-defined sequences that can
counteract the forces of homologous pairing under
experimental conditions. Trough the use of different
approaches, such as transgenic assays, several short
sequences from the Abd-B have been shown to be able
to mediate regulatory interactions over exceedingly
large distances (sometimes between different chromo-
somes). Two of these sequences are derived from the
Mcp [11], and the Fab-7 [12] regions. Both contain a
boundary and at least one PRE, and are able to medi-
ate long-distance regulatory interactions via the associ-
ation between homologous regions. In transgenic lines,
these sequences can interact with another copy inserted
somewhere else in the genome, or with their homolog-
ous sequence in the BX–C. These interactions between
distantly located copies usually result in silencing of
the reporter gene, or a gene next to the insertion site
of the transgene, although Mcp can also mediate posit-
ive regulatory interactions in exceptional cases [11].
However, these effects are observed only if at least one
copy of them is present in a transgenic insert, and the
significance of this high affinity pairing in the regula-
tion of the Abd-B is therefore unclear. Perhaps tight
homologous pairing between these sequences within
the BX-C plays a role in restricting the extent of loop-
ing-out domains.
Tethering elements
Deletion analysis of the Abd-B gene strongly suggests
the existence of a novel mechanism that tethers cis-
regulatory regions to the promoter-upstream region
[13]. It has been found that while Abd-B point muta-
tions do not complement the phenotype of an iab-7
deletion in A7, Abd-B alleles deleted for the promoter
region do complement iab-7 deletions in trans-hetero-
zygotes. The complementation is a result of the
action of the wild-type iab-7 on the wild-type Abd-B
in trans (Fig. 3). As this trans regulation is not detec-
ted when the somatic pairing of homolog chromo-
somes is disturbed by chromosomal rearrangements,
it represents a case of ‘transvection’. (The term
L. Sipos and H. Gyurkovics Long-distance interactions in Abd-B
FEBS Journal 272 (2005) 3253–3259 ª2005 FEBS 3255


























